Research Article, Vector Biol J Vol: 2 Issue: 2
Identification and Antibioresistance Characterisation of Culturable Bacteria in the Intestinal Microbiota of Mosquitoes
Yerbanga RS1,2*, Aminata Fofana1, Mathilde Gendrin3,4, Ibrahim Sangare5,6, Soufiane Sanou6, Soumeya Ouangraoua7, Somé AF1, Aly Drabo7, Jacques Simpore8, Thierry Lefèvre9,10, Anna Cohuet9, George Christophides3 and Ouédraogo JB1
1Institut de Recherche en Sciences de la Santé, Bobo Dioulasso , Burkina Faso
2Institut des Sciences et Techniques (INSTech Bobo), BP2779 Bobo Dioulasso, Burkina Faso
3Department of Life Sciences, Imperial College London, UK
4Microbiota of Insect Vectors group, Institut Pasteur de la Guyane, Cayenne, French Guiana
5Université Polytechnique de Bobo Dioulasso, Burkina Faso
6Centre Hospitalier Universitaire Sanou Sourou, Bobo Dioulasso, Burkina Faso
7Centre Muraz, Bobo-Dioulasso, Burkina Faso
8University of Ouagadougou, CERBA/LABIOGENE, Burkina Faso
9MIVEGEC (Maladies Infectieuses et Vecteurs: Ecologie, Génétique, Evolution et Contrôle), Montpellier, France
10CNRS UMR5290-Université de Montpellier, Montpellier, France
*Corresponding Author : Rakiswendé Serge Yerbanga
Institut de Recherche en Sciences de la Santé, Bobo Dioulasso, Burkina Faso
E-mail: stefanbio@yahoo.com.br, yrserge@yahoo.fr, h yrserge@yahoo.fr
Received: December 05, 2017 Accepted: December 18, 2017 Published: December 30, 2017
Citation: Yerbanga RS, Fofana A, Gendrin M, Sangare I, Sanou S, et al. (2017) Identification and Antibioresistance Characterisation of Culturable Bacteria in the Intestinal Microbiota of Mosquitoes. Vector Biol J 2:2. doi: 10.4172/2473-4810.1000123
Abstract
Background: The bacterial microbiota which colonize the mosquito midgut play an important role in vector-parasite interactions and consequently can modulate the level of malaria transmission. Their characterization may contribute to new control strategies of malaria transmission. However, these bacteria may also be eliminated in areas of high antibiotics usage. In this study, we identified paratransgenesis bacteria candidate in the gut of adults female Anopheles in Burkina Faso.
Methods: The gut of 73 semi-field mosquitoes and 28 laboratory-reared mosquitoes from two villages in Burkina Faso were analyzed by conventional in vitro culture techniques to isolate and identify bacteria of the microbiota. The gene 16S sequencing was used to confirm the presence of bacteria of paratransgenesis interest. Due to the effect of antibiotics on bacteria, we evaluated in vitro their susceptibility to antibiotics generally used for infectious diseases treatment. Results: In total, eleven genera of bacteria were identified: Pantoea, Sphingomonas, Escherichia, Micrococcus, Staphylococcus, Klebsiella, Serratia, Acinetobacter, Pseudomonas, Citrobacter, Asaia. Among these bacteria isolated, Asaia. sp and Pantoea. sp have already been reported as candidates for paratransgenesis. In addition, we observed pathogenic bacteria such as Escherichia coli, Klebsiella pneumonia and Pseudomonas luteola. Investigation of the correlation between the bacterial microbiota and malaria infection status showed that mosquitoes engorged with blood containing Plasmodium falciparum contained a higher bacterial load than non-blood fed mosquitoes. The antibiotic susceptibility test showed that Asaia, Pantoea and Serratia, previously proposed as paratransgenesis candidates, were susceptible to different antibiotics tested in contrast to Escherichia coli, which were resistant.
Discussion: Midgut analysis shows that the composition of the bacterial microbiota in wild field mosquitoes exhibits a large variability in contrast to laboratory-reared mosquitoes. The presence of genera already proposed as paratransgenesis candidates in previous studies among bacteria isolated in our study, suggested the possible implementation of this control strategy in Burkina Faso. Nevertheless, our data indicates that an in vivo verification of the stability of these bacteria is needed, as this strategy may be impaired by mass drug administration programs and antibiotic misuse.
Keywords: Escherichia coli; Klebsiella pneumonia;Pseudomonas luteola
Introduction
Malaria remains a serious health problem in developing African countries, causing 446000 deaths annually [1]. Plasmodium falciparum is the most prevalent species that causes malaria in sub-Saharan Africa and is responsible of most of deaths.Mainly the mosquito, Anopheles gambiae, transmits P. falciparum [2,3].
In recent years, scaling up of malaria control interventions has led to important progress in reducing the malaria burden in several countries. However, despite the large application of interventions such as insecticide-treated nets (ITNs) distribution, artemisinin-based combination therapy (ACT) and intermittent preventive therapy (ITP), malaria remains one of the greatest global health challenges. Emerging insecticide-resistant vectors and environmental issues related to the application of pesticidesin agriculture have necessitated the development of new control strategies with less environmental impacts or damage and higher efficacy. Paratransgenesis is one of the promising new strategies that use endogenous mosquito bacteria to interfere with the proliferation of malaria parasite and to block the transmission of malaria to human.
The mosquito gut accommodates a complex microbial community comprising diverse microorganisms, which are essential for various mosquito life traits, such as development and fecundity [4-6]. The protective role of Anopheles midgut bacteria against malaria infections has also been demonstrated by using antibiotic treatments to clear the gut microbiota, which resulted in enhancing Plasmodium infections [4,7]. The protection mechanism involves the direct interactions between bacteria and parasites and indirect interactions as the stimulation of the mosquito immune system by bacteria [4,8]. Whether natural or induced by genetic modification, the microbiota-induced refractoriness is regarded with growing interest in studies on malaria transmission [9-12]. So, it is necessary to characterize the normal midgut microbiota of mosquitoes and choose the best candidates for paratransgenesis strategy. Focus points of paratransgenesis applicability are the ease in genetic manipulation to induce refractoriness to Plasmodium, the efficiency in spreading into the mosquito population and safety. The increasing use of antibiotics and pesticides may affect the mosquito gut microbiota and also introduce a selection pressure for resistance in microorganism populations [13]. A recent study has shown that antibiotics present in ingested human blood reduced the bacterial load in the midgut of mosquitoes, which increases their susceptibility to the malaria parasite and its transmission [14]. In regard to the extension of antibiotic use, it is important to investigate the impact of antibiotics on bacteria isolated from the midgut of mosquitoes to evaluate their resistance. In terms of safety, an ideal paratransgenesis candidate should be sensitive to at least some antibiotics. However, quantifying its sensitivity to antibiotics of frequent use or distributed during mass drug administration programs will inform the elaboration of paratransgenesis agendas.
In the present study, we isolated culturable bacteria from the midgut of An. gambiae and An. coluzzii from two localities in Burkina Faso to identify potential candidates for paratransgenesis. Additionally, we evaluated the resistance of bacteria isolated to few antibiotics to assess the colonization success of target bacteria for paratransgenesis.
Materials and Methods
Ethic statement
All procedures involving human subjects used in this study were approved by the national and institutional ethical committee of Burkina Faso (statement N°2015-5-056/CERS and A02-2015/ CEIRES). The gametocyte carrier used in this study was enrolled as volunteers after his parents had signed a written informed consent form.
Larvae collection and mosquito rearing
Mosquito larvae were sampled in aquatic habitats at the L4 and pupae stages in Soumousso and Vallée du Kou, two villages situated respectively about 50 km and 30 km of Bobo-Dioulasso (Burkina Faso) using standard dipping techniques in August, a raining season. These localities were characterized by Moritz [15-17]. In each locality, breeding sites were inspected visually for the presence of larval stages. And, larvae were collected and kept in a water container for transportation to the insectary at the Institutde Recherche en Sciences de la Santé (IRSS) in Bobo-Dioulasso.
Anopheline larvae were identified morphologically; nonanopheline larvae and predators were removed. Larvae were kept in their original habitat water in a plastic bucket and the resulting pupae were collected daily for 2 days. Pupae were transferred to a plastic cup containing water from the breeding site, and the cup was placed in a cage for emergence. Adult mosquitoes were maintained in standard insectary conditions (26-27ºC, 65-85% humidity, and 12 h: 12 h light/ dark cycle) and provided with 5% sterile glucose solution.
Regarding laboratory-reared mosquitoes, the colony was maintained in insectary conditions. Adult females were fed on rabbit blood, then left to lay eggs on wet filter papers. Emerging larvae were fed with Tetramin® fish food and kept in clean water from a local spring. Some mosquitoes were fed on blood containing P. falciparum gametocytes to assess the effect of feeding on infectious blood on bacterial diversity.
Selection of gametocyte carriers and blood feeding
Children aged 5 to 12 years from Soumousso (school, health district), who showed no clinical symptoms of malaria were screened for presence of gametocytes in the blood. Thick smears were brought to the laboratory and stained with Giemsa and examined underlight microscope (1000X) to detect P. falciparum gametocytes. The next day, new thick and thin smears were performed on peripheral blood of the selected gametocyte-carrier to determine parasitemia at the time of the experiment. Approximately 4mL of venous blood were sampled into a heparinized tube. The mosquito infection was performed using an experimental device composed of several feeders kept at 37°C by warm water circulation. Each feeder was filled with 400 μL heparinized blood and closed with a Parafilm membrane, through which mosquitoes were allowed to feed for at least15-30 minutes. After removal of non-blood fed mosquitoes, they were distributed in different cages and reared under the climatic conditions in the insectary.
Sample selection
In this study, 101 individual midguts of mosquitoes from different localities were selected for bacterial characterization. We included a control group of 100 laboratory reared mosquitoes from the same locality and used F1 generation for the experimental infection in order to investigate bacteria-P. falciparum interaction. After blood feeding, 57 midguts dissected were subjected to infectivity analysis.
Isolation of the mosquito midgut bacteria
Mosquitoes were cold-anesthetized and all dissections were processed under clean conditions. Mosquitoes were surface sterilized with a 70% ethanol solution, rinsed three times in sterile phosphatebuffered saline (PBS) and dissected on ethanol-sterilised slides. Midguts were individually ground in 50 μl sterile PBS using a singleuse pestle and carcasses individually stored in 70% ethanol and maintained at -20°C for the determination of the mosquito species. 20 μL of each midgut homogenate was cultured in Brain-Heart infusion (BCC) medium composed of bovine brain and heart for 24 to 48hours. After incubation, each broth was sub-cultured on solid medium called Bromo-cresol-Purple (BCP) composed of meat extracts, peptones and agar to isolate colonies. Colonies with distinct morphologies, colours and margins were Gram-stained and sub-cultured into other media such as blood agar for diplococci, Chapman for cocci, Eosin Methylene Blue (EMB) and Cystine Lactose Electrolyte Deficient (CLED) including yeast extracts, peptones, L-cystine, electrolyte deficiency and agar for Gram-negative bacilli. After incubation, individual bacterial colonies were restreaked onto Müller Hinton medium (MH) composed of yeast and meat extracts and peptones to obtain pure bacterial isolates. Cultures were maintained at 37°C under aerobic conditions for 24 to 48 hours.
Identification of the bacterial colonies
Bacterial isolates were identified by characterizing their culturing conditions and biochemical characters using API20 galleries (Biomérieux) following the manufacturer’s instructions. Some colonies isolated from mosquito midguts were identified via 16S rDNA sequencing. The 16S rDNA gene was amplified by colony PCR using the Gotaq polymerase with the following primers: 16S-27F (TTAGAGTTTGATCCTGGCTCAGAG) and 16S-1391R (TACGGYTACCCTTGTTACGACTTC). 30 cycles (93ºC for 30s, 58ºC for 30s, 72ºC for 1min30s) were performed. The PCR product was checked on a 0.8% agarose gel and purified with a Qiagen PCR purification kit prior to Sanger sequencing (Beckman Coulter Genomics) using the same primers.
Antibiotics susceptibility testing
In total of 9 antibiotics: Ampicillin (10 μg/mL), Amoxicillin/ Clavulanic acid (20 μg +10 μg/mL), Ceftriaxone (30 μg/mL), Gentamicin (10 μg/mL), Norfloxacin (10 μg/mL), Co-trimoxazole (25 μg/mL), Erythromycin (15 μg/mL), Oxacillin (1 μg/mL) and Penicillin (10 μg/mL) according to Clinical and Laboratory Standards Institute recommendation in bacterial infections (Ca-SFM 2014) were selected to perform the sensitivity test. The susceptibility of bacteria to antibiotics was evaluating using Kirby-Bauer disk diffusion method on Müller Hinton agar.
Statistical analyses
All statistical analyses were performed in R (version 3.3.0). Logistic regression by Generalized Linear Models (GLM, binomial errors, logit link) was used to investigate the effect of mosquito origin (VK, Soumousso, and laboratory), mosquito species and interaction on the probability to obtain positive bacterial culture from mosquito midgut. We also tested the effect of mosquito engorgement on infectious blood meal and interaction with mosquito origin using a GLM binomial. Multiple pair-wise comparisons were performed using the function glht of the multcomp package.
Results
Vector population
A total of 101 An. gambiae mosquitoes were included in this study. 73 were collected at the larval stage in the field and reared until they reached the adult stage in the insectary. Among these, 20 originated from Vallée du Kou, an irrigated rice field area (17 An. coluzzii and 3 An. gambiae), 53 originated from Soumousso (11 An. coluzzii and 42 An. gambiae). 28 were laboratory-reared mosquitoes (7 An. coluzzii and 21An. gambiae).
Composition of the bacteria microbiota of the mosquito midgut
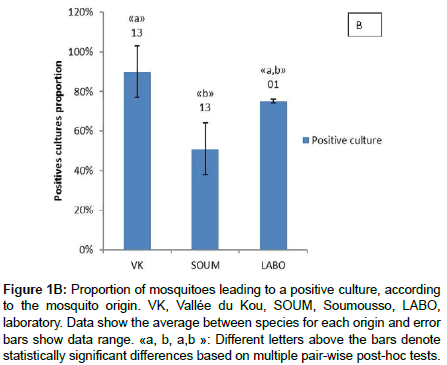
Bacterial culture on different media was successful in 65% (66/101) of the midguts, and 38% (25/66) of the isolates were successfully identified. A total of 12 bacterial genera (Table 1) were isolated and Gram-negative bacteria were prevailing (68%, 17/25). There was a significant site effect on the probability of cultivating bacteria from mosquito midgut (LRT X22=12.397, P=0.002). Mosquitoes of Vallée du Kou exhibited more positive culture than mosquitoes of Soumousso (90% versus 51%). Positive cultures were obtained from 75% of laboratory mosquitoes and this proportion was not statistically different from Soumousso and VK mosquitoes (Figure 1B). However, no significant difference (71% versus 59%; LRT X21=1.2, P=0.27) was found between An. coluzzii and An. gambiae (Figure 1A). Finally, there was no mosquito species by locality interaction (LRT X22= 2.1, P=0.34).
Figure 1B: Proportion of mosquitoes leading to a positive culture, according to the mosquito origin. VK, Vallée du Kou, SOUM, Soumousso, LABO, laboratory. Data show the average between species for each origin and error bars show data range. «a, b, a,b »: Different letters above the bars denote statistically significant differences based on multiple pair-wise post-hoc tests.
| Field mosquitos | Laboratory reared mosquitos | Total | ||||
|---|---|---|---|---|---|---|
| Habitat | Soumousso | Vallée du Kou | IRSS insectary | |||
| Mosquito species | An. coluzzii | An.gambiae | An.coluzzii | An.coluzzii | An. gambiae | |
| Bacteria species | ||||||
| Pantoea sp. | 0 | 1 | 0 | 1 | 1 | 3 (13. 6%) |
| Sphingomonas paucimobilis | 1 | 0 | 0 | 0 | 0 | 1 (4.5%) |
| Escherichia. coli | 0 | 1 | 0 | 0 | 0 | 1 (4.5%) |
| Negative-coagulase Staphylococci | 0 | 4 | 0 | 1 | 0 | 5 (22.7%) |
| Micrococcus sp | 0 | 1 | 0 | 0 | 0 | 1 (4.5%) |
| Serratia ficaria | 1 | 0 | 0 | 0 | 1 (4.5%) | |
| Staphylococcus epidermidis | 0 | 0 | 1 | 0 | 0 | 1 (4.5%) |
| Klebsiella pneumoniae | 0 | 0 | 1 | 0 | 0 | 1 (4.5%) |
| Acinetobacter lwoffii | 0 | 0 | 0 | 0 | 2 | 2 (9.1%) |
| Pseudomonas luteola | 0 | 0 | 0 | 0 | 1 | 1 (4.5%) |
| Citrobacter freundii | 1 | 0 | 0 | 3 | 0 | 4 (18 .2%) |
| Pantoea sp. 2 | 0 | 0 | 0 | 1 | 0 | 1 (4.5%) |
| TOTAL | 3 | 7 | 2 | 6 | 4 | 22 (100%) |
Table 1: Bacteria composition of field and laboratory reared mosquitoes by biochemical identification.
In field mosquitoes, we identified 12 bacteria species. Enterobacteriaceae and Staphylococcaceae were the dominant families. Among which, coagulase-negative Staphylococci was the most common (80%). For example, Pantoea sp., Citrobacter freundii and Acinetobacter lwoffii were isolated more than one mosquito (Table 1).
To get a broader view of the bacterial microbiota, 8 colonies isolated from individual mosquitoes of the three localities were identified using 16S gene sequencing. We identified 5 Gram-negative bacteria belonging to the phyla of Proteobacteria and Actinobacteria (Table 2).
| Phylum | Class | Family | Species | Accession numbers NCBI | Max id. | Origin of bacterial isolates/mosquito species | ||
|---|---|---|---|---|---|---|---|---|
| Soumousso | Vallée du Kou | Lab | ||||||
| An. gambiae | An. gambiae | An.coluzzii | ||||||
| Proteobacteria | γ-proteobacteria | Enterobacteriaceae | Pantoeadispersa | HQ443235.1 | 97% | 1 | 0 | 0 |
| Proteobacteria | γ-proteobacteria | Aeromonadaceae | Aeromonasjandaei | HQ683971.1 | 99% | 1 | 0 | 0 |
| Proteobacteria | β-proteobacteria | Burkholdheriaceae | Burkholderia gladioli | CP002600.1 | 95% | 0 | 1 | 0 |
| Proteobacteria | α-proteobacteria | Acetobacteraceae | Asaia sp. | FN814295.1 | 99% | 0 | 0 | 1 |
| Actinobacteria | Actinobacteria | Microbacteriaceae | Microbacterium sp. | JQ418292.1 | 100% | 0 | 0 | 1 |
Table 2: Isolated bacterial by sequencing of 16S genus.
Composition of the bacterial microbiota following exposition to P. falciparum-infected blood
To characterize the bacterial microbiota in the midgut of mosquitoes infected by malaria parasites, mosquitoes of different origins (laboratory or F1 generation mosquitoes) were offered blood infected by P.falciparum gametocytes (104/μl) with 11.7% of haemoglobin). On day 7 after the blood meal, the observation of the control midgut showed a prevalence of oocystes infected mosquitoes of 61 ± 12% with an average of 3.8 ± 0.58 oocystes per midgut. Midguts of P.falciparum-exposed mosquitoes were more likely to produce positive bacterial culture than midgut of non-engorged mosquitoes (77% versus 55%; LRT X22=5, P=0.025). However, there was a significant mosquito origin by P.falciparum exposure interaction indicating that this effect was not consistent across all mosquito origin (LRT X22=8, P=0.017). In particular, engorged mosquitoes from Soumousso displayed less positive cultures than laboratory and VK mosquitoes (Figure 2). Of note, the bacterial microbiota of the midgut was mainly composed of Firmicutes belonging to the family of Staphylococcaceae (Staphylococcus epidermidis, coagulase-negativeStaphylococci) and Proteobacteria belonging to the Enterobacteriaceaefamily (Klebsiellapneumoniae, Citrobacterfreundii, Acinetobacterlwoffii, Pantoea.spandPantoea sp2) (Table 1).
Figure 2: Proportion of mosquitoes leading to a positive culture, according to the mosquito blood feeding status. Engorged mosquitoes were fed with P. falciparum infected blood 7 days prior to sampling. VK, Vallée du Kou, SOUM, Soumousso, LABO, laboratory. Data show the average between species for each origin and error bars show data range.
Antibiotic resistance test of the isolated bacteria
We assessed the impact of antibiotics on the bacterial isolates to verify their resistance. All the bacteria strains were tested to 3 different antibiotics commonly used in Burkina Faso (Table 2) resulting an important sensitivity (16/20). Asaia sp. the bacteria target in paratransgenesis [18,19] were totally inhibited (100%) by ampicillin, Amoxicillin/Clavulanic acid, ceftriaxone, gentamicin, norfloxacin and cotrimoxazole. In contrast, Escherichia coli were resistant (100%) to all antibiotics tested. Pantoea strains isolated from different mosquitoes as presented different profile of sensitivity to different antibiotics tested. Of note, Serratia strain, another probably potential candidate of paratransgenesis [20,21] was resistant to Ampicillin, Gentamicin, Norfloxacin and Cotrimoxazol. However, it was sensitive to Amoxicillin/Clavulanic acid and ceftriaxone (Table 2).
Discussion
The overall aim of this study was to assess the paratransgenesis implementation on Burkina Faso. We therefore investigated the normal midgut microbiota of adult female Anopheles mosquitoes from different origins in Burkina Faso (Soumousso,Vallée du Kou and laboratory). Our data showed that the midgut of mosquitoes accommodated an important population of the bacterial microbiota among which some bacterial genera already considered as potential paratransgenesis candidates [18,21,22]. We also tested the antibioticresistance of bacteria isolated in order to test their survival capacity in the midgut.
Exploration of individual midgut of mosquitoes from different natural environments using culture dependent method was successful in 65% samples. So, we noticed that mosquitoes of Vallée du Kou accommodated most bacteria in relation to Soumousso (90 vs 51) indicating an environment impact on bacteria diversity [23]. In contrary, Boissiere [24] observed a higher similarity between adults mosquitoes derived from the same pond than mosquitoes derived from different breeding site [24]. As previously reported, we also observed an important bacterial diversity in field mosquitoes as in laboratory-reared mosquitoes which practically presented the same bacteria species [24]. According to previous studies [25-27], laboratory mosquitoes are raised in a controlled environment where they come in contact with a less diverse set of bacteria. Unlike laboratory mosquitoes, field mosquitoes feed on nectar and fruit from bacteria-harbouring plants and larvae ingest microorganisms contained in the breeding sites that were used in their development.
Among the positive cultures, only 38% essentially Gram-negative bacteria were identified. As previous studies [4,24,28,29], Gammaproteobacteria class were dominant. Through determination of the analytical profile index of our isolates, we identified enteric bacteria such as Serratia ficaria, Escherichia coli and Aeromonas jadaei commonly isolated in the human gut. These genera of bacteria are some of the most commonly harboured by Anopheles mosquitoes suggesting that they may adapt easily to the midgut environment and thus may be interesting for paratransgenesis purposes [11]. Nevertheless, these bacteria are often isolated in nosocomial infections. So, further investigations on these strains will be interesting to verify if they can be considered as good paratransgenesis candidates or if their spread may represent a risk for human health, ruling out the development of a paratransgenic candidate from our strain of Escherichia coli, which is resistant to every antibiotic tested.
We also identified by16S gene sequencing some of the colonies that we had not found by determining the analytical profile index. Burkholderia gladioli were found in the midgut of wild strains of mosquitoes from the Vallée du Kou, a rice field area. Burkholderia gladioli is a phytopathogenic species that causes rot of orchids, onion and iris [30]. In rice fields, this bacterium inhibits the proliferation of other species such as Burkholderia glumae and Burkholderia plantarii which cause rot of grains and rice seedlings [20,30,31]. This symbiont is capable of protect insects against entomopathogenic fungi [32,33]. The presence of this species could be the result of a colonisation from the breeding site [10] also isolated the genus Burkholderia. sp mosquitoes from Burkina Faso [10]. The effect of Burkholderia symbiont on malaria vectors remains unknown to date [24] but it remain a dangerous microbiota for plants.
Asaia sp. and Microbacterium sp. were identified of the midgut of laboratory mosquitoes. Asaia sp is anacetic acid bacterium that can be contracted either by oral route during feeding in the larval habitats or from flower nectar [10]. This bacterium is known to have all the required ecological characteristics that make it the best candidate available to date for the development of paratransgenesis. It may be transmitted horizontally through mating in adult mosquitoes, vertically transmitted from mother to offspring [10,19] indicating that it could spread rapidly in wild mosquito populations.
However, our data show that this bacterium is sensitive to all antibiotics commonly used in Burkina Faso, suggesting that it could be affected by human blood ingested by mosquitoes. Indeed, the presence of antibiotics in a blood meal decreases the growth of bacterial gut and also modifies the composition of bacteria community [14]. Other interesting bacteria for the paratransgenesis such as Serratia, and Pantoea genera were identified [21], and have shown variability in their susceptibility to the different antibiotics. We noticed that some of these bacteria strains were inhibited to a greater extent after treatment with co-trimoxazole, gentamicin and norfloxacin. Our results suggest that a high use of antibiotics may negatively affect the paratransgenesis implementation in Burkina Faso.
In contrast to Asaia, Escherichia presented a great resistance to different antibiotics tested). This resistance could be explaining by the spread of resistant bacteria in the environment after a non-controlled human waste. Although no mosquito microbiota transmitted to humans during the bite has yet been reported, further studies may determine if the presence of this multidrug resistant bacterium in the mosquito microbiota is a risk for human health.
Comparing the susceptibility of the bacterial microbiota according to mosquito species, we did not identify any significant difference (P=0.27) between An. coluzzii and An. gambiae. However, variability of bacteria species was noticed within An. coluzzii and An. gambiae (Tables 1 and 2). Although the origin of the microbiota is still largely unclear, the inter individual variability of mosquitoes reared on sterile food as adults from field-collected larvae is partly explained by the larval habitat [34]. A few bacterial taxa are known to be transstadially transmitted in Anopheles sp. However in Aedes aegypti Coon et al. 2014 showed that a variety of bacteria detected in larvae are transmitted to adults [35]. Nevertheless, as the microbiota of mosquitoes collected at the adult stage is not explained by the geographical origin [28], the adult microbiota is likely to be also highly influenced by ingestion of bacteria from the adult environment.
Previous studies revealed a compromising effect of bacterial infection on malaria [5,20,34]. We therefore investigated the coinfection of bacteria and P. falciparum. Thus, mosquitoes of laboratory and field were exposed to blood meal of the same gametocyte carrier. After 7 days followed blood feeding, mosquitoes were indeed infected (4 oocystes/mosquito). We observed in the batch of engorged mosquitoes a significant proliferation of bacteria colony from midgut of mosquitoes engorged (P=0.025) in contrast to mosquitoes none engorged. We also compared cultures of mosquito guts according to locality and P. falciparum infection status. The results showed that mosquitoes engorged with blood infected from Vallée du Kou and laboratory harboured more bacteria than counterparts from Soumousso and that Enterobacteriacae were the dominant family (60%). We think that diversity of the bacterial microbiota could depend on the origin of mosquito and of its P. falciparum infection status. A recent study suggested that an interaction Enterobacteriacae-P. falciparum may promote successful development of either [24]. Other studies [4,9] associated the mosquito susceptibility to malaria parasite on bacteria absence but not a relationship between mosquito co-infection bacteria-P. falciparum and their origin. Mosquito engorged and non-engorged presented overall similar bacteria species diversity. However, our analysis using cultures and analytical profile index was limited due to the need of bacterial proliferation for their identification. In the laboratory-reared mosquitoes, most bacteria were isolated from An. coluzzii after an infected blood meal through membrane feeding assay while we observed a weak colonization in the uninfected mosquitoes. Wang and colleagues followed the evolution of the microbiota composition of a colony of laboratory An. gambiae during its development cycle, and observed a decrease in the diversity of the bacterial microbiota in the adult mosquitoes after a blood meal [36].
Conclusion
This characterization of the mosquito gut microbiota is a first step toward the selection of paratransgenesis candidates to control malaria transmission in Burkina Faso. Nevertheless, it will be interesting to widen the sampling collection and screening mosquito midgut through sequencing of the bacterial microbiota in Burkina Faso. The results indicate that, despite the presence of paratransgenic candidates Asaia sp., Pantoea sp. and Serratia sp. among the isolated bacteria, the application of this strategy should consider the impact of antibiotics on the bacterial microbiota. So, the misuse of antibiotics and mass drugs administration programs could potentially affect genetic modification or result in the elimination of the target bacteria.
References
- WHO (2016) World malaria report.
- Della TA, Tu Z, Petrarca V (2005) On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol 35: 755-769.
- Lawniczak M, Emrich SJ, Holloway AK, Regier AP, Olson M, et al. (2010) Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330: 512-514.
- Dong Y, Manfredini F, Dimopoulos G (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5.
- Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC (1996) Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am J Trop Med Hyg: 54: 214-218.
- Chouaia B, Rossi P, Epis S, Mosca M, Ricci I, et al. (2012) Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol 8.
- Beier MS, Pumpuni CB, Beier JC, Davis JR (1994) Effects of paraaminobenzoic acid, insulin, and gentamicin on Plasmodium falciparum development in anopheline mosquitoes (Diptera: Culicidae). J Med Entomol 31: 561-565.
- Meister S, Agianian B, Turlure F, Relogio A, Morlais I, et al. (2009) Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog 5.
- Cirimotich CM, Dong Y, Clayton AMSL, Sandiford SJA, Mulenga M, et al. (2011) Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science: 332 (6031): 6855-6038.
- Favia G, Ricci I, Damiani C, Raddadi N, Crotti E (2007) Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci 104: 9047-9051.
- Gendrin M, Kristophides GC (2013) The Anopheles Mosquito Microbiota and Their Impact on Pathogen Transmission. Medicine » Infectious Diseases » "Anopheles mosquitoes - New insights into malaria vectors.
- Wang S, Ghosh AK, Bongio N, Stebbings KA, Lampe DJ, et al. (2012) Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. PNAS 109.
- Hill CA, Kafatos FC, Stansfield SK, Collins FH (2005) Arthropod-borne diseases: vector control in the genomics era. Nat Rev Microbiol: 262-268.
- Gendrin M, Rodgers FH, Yerbanga RS, Ouedraogo JB, Basanez MG, et al. (2015) Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nature communication 7.
- Moritz Buck, Louise KJ, Nilsson, Carl Brunius, Dabiré RK, Richard Hopkins (2016) Bacterial associations reveal spatial population dynamics in Anopheles gambiae mosquitoes. Scientific report 6:22806
- Baldet, Diabate A, Guiguemde RT (2003) La transmission du paludisme dans la zone rizicole de la vallée du Kou (Bama), (Burkina Faso 55-60
- Robert V, Gazin P, Boudin C, Molez JF, Ouedraogo V, Carneval P (1985) La transmission du paludisme en zone de savane arborée et en zone rizicole des environs de Bobo-Dioulasso, Burkina Faso. Méd Trop 2: 201-214.
- Favia G, Ricci I, Marzorati M, Negri I, Alma A et al. (2008) Bacteria of the genus Asaia: a potential paratransgenic weapon against malaria. Adv Exp Med Biol 627: 49-59.
- Damiani C, Ricci I, Crotti E, Rossi P, Rizzi A (2010) Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb Ecol 60: 644-654.
- Gonzalez CL, Santillan F, Rodriguez MH, Mendez D, Hernandez AJE (2003) Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol 40: 371-374.
- Wang S, Dos-Santos ALA, Huang W, Liu KC, Oshaghi MA, et al. (2017) Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 1399-1402.
- Sibao Wang, Marcelo Jacobs-Lorena (2013) Genetic approaches to interfere with malaria transmission by vector mosquitoes. Trends Biotechnol 31: 185-193.
- Chavshin AR, Oshaghi MA, Vatandoost H, Pourmand MR et al. (2014) Isolation and identification of culturable bacteria from wild Anopheles culicifacies, a first step in a paratransgenesis approach. Parasit Vectors 7: 419.
- Boissiere A, Tchioffo MT, Bachar D, Abate L, Marie A, et al. (2012) Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog: 8-5.
- Gary R, Foster WA (2004) Anopheles gambiae feeding and survival on honeydew and extra-floral nectar of peridomestic plants. Med Vet Entomol 18: 102-107.
- Gouagna LC, Poueme RS, Dabire KR, Ouedraogo JB, Fontenille D, et al. ( 2010) Patterns of sugar feeding and host plant preferences in adult males of An. gambiae (Diptera: Culicidae). J vector ecology 35: 267-276.
- Chavshin AR, Oshaghi MA, Vatandoost H, Pourmand MR, et al. (2012) Identification of bacterial microflora in the midgut of the larvae and adult of wild caught Anopheles stephensi: A step toward finding suitable paratransgenesis candidates. Acta Trop 121: 129-134.
- Osei-PJ, Mbogo CM, Palmer WJ, Jiggins FM (2012) Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Molecular Ecology 21: 5138-5150.
- Tchioffo MT, Boissiere A, Churcher TS, Abate L, Gimonneau G, et al. (2013) Modulation of Malaria Infection in Anopheles gambiae Mosquitoes Exposed to Natural Midgut Bacteria. PLoS ONE 12.
- Keith L (2005) Isolation and characterization of Burkholderia gladioli from Orchids in Hawaii. Plant Disease 89: 1273-1278.
- Gonzalez CF, Venturi V, Engledow AS (2007) The phytopathogenic Burkholderia. In: Burkholderia, molecular microbiology and genomics (Coenye T. and Vandamme P., eds). Horizon Bioscience, Norfolk, United Kingdom: 153-176.
- Kikuchi Y, Hosokawa T, Fukatsu T (2007) Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol 73: 4308-4316.
- Santos AV, Dillon RJ, Dillon VM, Reynolds SE, Samuels RI (2004) Occurrence of the antibiotic producing bacteriumBurkholderiasp. in colonies of the leafcutting antAtta sexdens rubropilosa. FEMS Microbiol Lett 239: 319-323.
- Straif S, Mbogo CN, Toure AM, Walker ED, Kaufman M, et al. (1998) Midgut bacteria in Anopheles gambiae and A. funestus (Diptera: Culicidae) from Kenya and Mali. J Med Entomol 35: 222-226.
- Coons KL, Vogel KJ, Brown MR, Strand MR (2014) Mosquitoes rely on their gut microbiota for development. Mol Ecol 23: 2727-2739.
- Wang Y, Gilbreath 3rd TM, Kukutla P, Yan G, Xu J (2011) Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi