Research Article, J Virol Antivir Res Vol: 8 Issue: 1
Estimation of the Role of Apolipoprotein E Genotypes in Patient Infected with Hepatitis C Virus and Treatment with Sovaldi Drug
El-Adly AM1*, Rashed LA2, Yehia HM2, Wardany AA1, Shikhoun MH3 and Abdul-Raouf UM1
1Botany and Microbiology Department, Faculty of Science, Al-Azhar University, 71524 Assiut, Egypt
2Faculty of Medicine, Al-Kaser Al-Any Hospital, Cairo University, Cairo, Egypt
3Analyses and Laboratories Department, Higher Technological Institute of Applied Health Sciences in Sohag, Ministry of Higher Education, Cairo, Egypt
*Corresponding Author: El-Adly AM Botany & Microbiology Department, Faculty of Science, Al-Azhar University, 71524 Assiut, Egypt, Tel: 01147787828; E-mail: ahmedeladly.ast@azhar.edu.eg
Received: January 18, 2019 Accepted: April 16, 2019 Published: April 22, 2019
Citation: El-Adly AM, Rashed LA, Yehia HM, Wardany AA, Shikhoun MH, et al. (2019) Estimation of the Role of Apolipoprotein E Genotypes in Patient Infected with Hepatitis C Virus and Treatment with Sovaldi Drug. J Virol Antivir Res 8:1.
Abstract
Background: Some single-nucleotide polymorphisms (SNPs) in lipid organizers such as apolipoproteins and cell surface molecules for hepatitis C virus (HCV) entry into hepatocytes are related to HCV infection. Silencing apoE expression resulted in marked inhibition of infectious particle production without affecting. Aim: This study aimed to investigate the effect of apoE genotype on the response to HCV treatment with sovaldi drug at Sohag governorate, Egypt. Method: Randomly, we selected 285 patients infected with hepatitis C virus from males and females with different age groups. All patients selected from Sohag governorate and treated with sovaldi drug for knowledge of the responder and non-responder for treatment by using RT-PCR technology. Results: Our results showed that out of 285 patients were infected with hepatitis C virus and treated with sovaldi drug, 258/285 (90.5%) patients were responded to treatment, and 27/285 (9.5%) patients non-responded to treatment. Out 285 patients were infected with hepatitis C virus and treated with sovaldi drug, we selected 50 patients (25 responded to sovaldi treatment and 25 non-responded to treatment) for other investigation. All patients aged between 30 to 55 years, where 13 (26%) cases of females and 37 (74%) cases of males. Our investigation showed that, in the case of respond to the treatment of Sovaldi drug, there are three genotypes of apoE ε3/ε2, ε3/ε3and ε3/ε4, which are 8 (32%), 2 (8%) and 15 (60%) respectively. In the case of non-response, there are 3 (12%) cases with ε3/ε2, 14 (56%) cases with ε3/ε3 and 8 (32%) cases with ε3/ ε4. The concentration apoE increases in the case of infection with HCV and when the non-respond to therapy, while decreased when the respond to therapy. Conclusion: This study shows the role of apolipoprotein E polymorphism on the outcome of the HCV infection after treatment with sovaldi drug.
Keywords: Hepatitis C virus; Genotypes; Sovaldi; Sohag; Apolipoprotein E
Keywords
Hepatitis C virus; Genotypes; Sovaldi; Sohag; Apolipoprotein E
Abbreviations
ELIZA: Enzyme-Linked Immunosorbent Assays; HCV: Hepatitis C Virus; PCR: Polymerase Chain Reaction; RNA: Ribonucleic Acid; RT-PCR: Reverse Ranscriptase PCR; ApoE: Apolipoprotein E; VLDL: Very-Low-Density Lipoprotein; HCC: Hepato-Cellular Carcinoma; siRNA: small interfering RNA
Introduction
Hepatitis C virus (HCV) infects about 185 million people worldwide [1]. HCV infection typically causes chronic liver disease and can lead to liver cirrhosis and hepatocellular carcinoma (HCC) [2]. HCV is an enveloped single-stranded RNA virus of positive polarity that is a member of the genus Hepacivirus in the family Flaviviridae [3,4]. Accumulating studies have revealed that the HCV life cycle is closely related to host lipid metabolism [5,6]. Apolipoproteins are proteins on the surface of lipoprotein particles and establish the structure of lipoproteins [7]. ApoE is defined as a polymorphic protein arising from three alleles [8]. The human apoE gene was widely studied and described to be located on chromosome 19, closely linked to the apo C-I/C-II gene complex [9]. The three major alleles, termed Epsilon-2, Epsilon-3 and Epsilon-4 have been reported [10]. The apoE-ε2, apoE-ε3 and apoE-ε4 protein isoforms, corresponding products of these alleles, differ only by a single amino acid at two residues. Where ApoE-ε2 contains cysteine at two residue 112 and 158 and ApoE-ε4 has arginine at both positions while apoE-ε3 has cysteine at residue 112 and arginine at residue 158 [11]. Less than 0.1% of the population has additionally, two minor alleles of the gene, ε1 and ε5. Three homozygous (ε2/ε2, ε3/ε3, ε4/ε4) and three heterozygous (ε2/ε3, ε2/ε4, ε3/ε4) genotypes will be determined by these three major alleles [12]. The main functions of the apoE protein are the transport of lipids around the body and the repair of damaged tissue [13]. ApoE is also a key determinant of the outcome of infection, with possession of certain alleles conferring protection against, damage caused by several infectious agents, including HIV [14] and HCV, the ApoE-ε4 allele confers protection against severe liver disease [15]. An important feature of the hepatocyte is its key role in lipid metabolism. Increasing evidence suggests that the HCV life cycle and hepatocyte lipid metabolism pathways are closely linked. Membrane vesicles containing the HCV replication complex have been shown to be highly enriched in proteins required for very-lowdensity lipoprotein (VLDL) assembly, including apolipoprotein B (apoB), apolipoprotein E (apoE), and microsomal triglyceride transfer protein [16,17]. Several lines of evidence suggest that the infection cycle of HCV might also involve lipoproteins and apolipoproteins. HCV is associated with lipoprotein particles in serum [18], and entry might involve LDLRs: COS-7 cells transfected with the human gene for LDLR bind more HCV particles than untransfected cells, and antibodies that block binding of ligands to LDLRs also block binding of HCV to cells[19, 20]. Polymorphisms in the genes encoding some apolipoproteins have been described that give rise to distinct forms of the protein with different biologic properties, such polymorphisms might contribute to the different outcomes of HCV infection [21]. In our study we investigated the relationship between apolipoproteins E genotype and their concentrations with response to Sovaldi drug. We found a strong relationship in the response to treatment with sovaldi drug with the infection of hepatitis C virus and apolipoprotein E genotypes. This fact shows that lipid metabolism plays an important role in the life cycle of hepatitis C virus. These results are intended to develop new treatments for chronic hepatitis C virus.
Materials and Methods
Ethics statement
Our study was performed randomly on 50 patients infected with Hepatitis C Virus without knowing apolipoprotein E genotype. All samples were collected from sohag hospital, Egypt during the period from November till July 2017, approved by patients and sohag hospital governorate. Blood samples were collected for different analysis at the time of routine clinic attendance. All patients included in our study were treated with Sovaldi drug for chronic HCV infection, informed written consent was obtained from each patient included in the study. Treatment regimеns and guidelines for enrollment with the national treatment programme were announced by the Ministry of Health, approved by patients and Sohag Hospital governorate.
Collection and processing of blood samples
We collected 285 blood samples patients infected with Hepatitis C virus, anti-HCV antibodies were tested by enzyme-linked immunosorbent assays (ELIZA) Technique and confirmed HCV RNA by the reverse transcriptase polymerase chain reaction (RTPCR). All patients included in our study were treated with sovaldi drug. After three months of treatment with sovaldi drug. Randomly, Out 285 patients were infected with hepatitis C virus and treated with sovaldi drug, we selected 50 patients for other investigation, 25/50 (50%) responded to sovaldi treatment, while others did not responded to treatment. The study was conducted on 50 known HCV-infected patients, recruitment of patients was random, and in the absence of knowledge of apoE genotype. For the HCV-RNA positive patients, blood samples were collected for different analysis at the time of routine clinic attendance. All patients included in our study were treated with Sovaldi drug. ApoE genotypes were determined by a modification of the PCR-based method. All blood samples were collected through the venous penetration using isopropyl alcohol at a concentration of 70% with 1% Iodine concentration and leaving the intravenous position for a minute to dry. All precautions were taken to avoid contamination of the site. Five milliliters of blood was collected using a stеrile syringe and needle and dispеnsed into clean plastic. The blood samples were centrifuged at 4000 rpm for 10 minutes, and the serum obtained was stored at -70ºC. Blood samples were collected for different analysis at the time of routine clinic attеndance. Our study was conducted on two different groups of pateints: the first group, which responded to sovaldi drug (n=25) (anti-HCV positive, HCVRNA negative patients) and the second group, which did not respond to sovaldi drug ((n=25) (anti–HCV-positive, HCV-RNA positive patients).
Serological assay
Detection of anti-HCV antibodies: All patients included in our study were revealed for anti-HCV antibodies by third-generation enzyme-linked immunosorbent assays [ELISA; BioKit-bioеlisa HCV 4.0]. The wells of microplate wеre coated with recombinant antigеns representing epitopes of HCV: core NS3, NS4 and NS5. The concentration of color is proportional to HCV antibodies concentration in the samplе.
Molecular assay
RNA isolation and RT-PCR: The presence of the viral gеnomе in sеrum was tеstеd by the reverse transcriptase polymerase chain reaction (RT-PCR) using the Amplicon method (Basеl, Switzerland). Viral RNA was extracted using thе viral RNA minutes kit according to thе manufacturеrs’ instructions by using spin column protocol (Qiagen, Hilden, and Gеrmany). The first strand complementary DNA (cDNA) was synthesized. Initial denaturation was performed at 95°C for 5 minutes. Polymerase chain reaction amplification was carried out at 94°C for 1 minute, 57°C (annealing temperature) for 1 minute, and 72°C for 1 minute for a total of 40 cyclеs and final еxtension at 72°C for 7 minutes. The primer sequences were used as follows: forward primеr was 5′CGCGCGACTAGGAAGACTTC3′ and reverse primer was 5′ACCCTCGTTTCCGTACAGAG 3′.
Electrophoresis agarose gel detection: The PCR-HCV products were detected by 1.5% agarose gel еlectrophoresis stained with ethidium bromide and spotted under ultraviolet light reactions. There are two treatment systems.
Pegylated interferon (pеg-INF)+ribavirin+sofosbuvir for 3 months.
Sofosbuvir+ribavirin for 6 months (for patients who are allowed to INF).
Apolipoprotein genotyping
DNA extraction from PBMCs: The method used for extraction of total DNA from lymphocyte for the detection of ApoE was done using extraction Kit, QIAamp DNA Mini and Blood for DNA purification from buffy coat spin column protocol purchased from QIAGEN. For DNA extraction, 20 μl of QIAGEN protease were pipette into 1.5 ml tube. Then, we added 200 μl of the sample to the tube, added 200 μl buffer AL and incubated at 56°C for 10 minutes. After short centrifugation to the tube, 200 μl of ethanol (96%) were added and mixed for 15 seconds. The mixture was applied to the QIAamp Mini spin column (in a 2 ml collection tube) and centrifuged at 8000 rpm for 1 minute. The QIAamp Mini spin column was placed in a clean 2 ml collection tube. Then, we added 500 μl of buffer AW1 to column, centrifuged at 8000 rpm for 1 minute and added 500 μl buffer AW2 to the column then, centrifuged of 14000 rpm for minutes. The QIAamp Mini spin column was placed in a new 1.5 ml micro centrifuge tube, then, we added 200 μl of buffer AE, incubated at room temperature for 1 minute and centrifuged at 8000 rpm for 1 minute.
ApoE genotypes determination
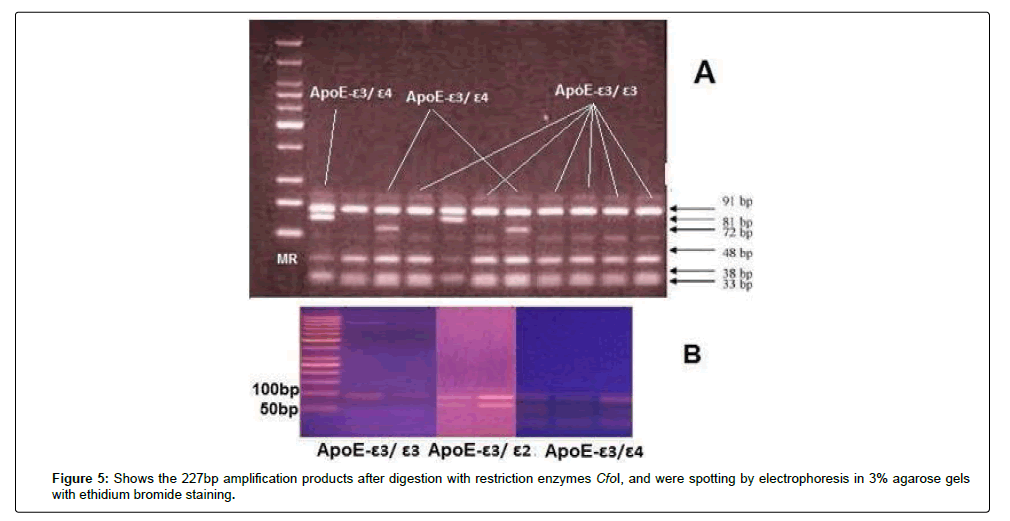
ApoE genotypes were determined according to previously published methods by Wenham; DNA was prepared from buffy coat according to the manufacturers’ instructions by using the QIAamp DNA mini kit. ApoE genotypes were determined by variation of the PCR-based method of Wenham. The sequence of the 2 primers, ApoE sense primer: 5’-TCCAAGGAGCTGCAG GCGGCGCA-3’ and ApoE antisense primer: 5`-ACA GAA TTC GCC CCG GCC TGG TAC ACT GCC A-3`. In the PCR, 5 μl of genomic DNA extracted from lymphocytes were added to 25 pmol of each primer and 25 μl of master mix and completed to 50 μl total volume by using deionized water. In the thermal reactor, an initial denaturation at 94ºC for 5 minutes followed by 40 cycles of denaturation at 94ºC for 30 seconds, annealing at 65ºC for 30 seconds, extension at 72ºC for 1.5 minutes and final extension at 72ºC for 10 minutes. The 227-basepair (bp) amplification products were digested for 2 hours at 37°C with restriction enzymes CfoI, then, spotted by electrophoresis in 3% agarose gels with ethidium bromide staining.
Detection
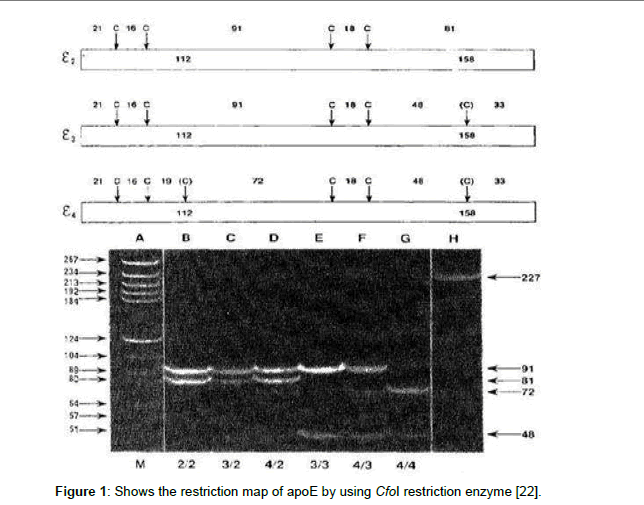
The PCR amplicon was detected by 3% agrose gel electrophoresis containing 10 μl of ethidium bromide (10 mg/ml), using a miniprep electrophoresis chamber (Pharmacia, LKB). The electrophoresis tank was filled with tris-acetate EDTA buffer. 10 μl of PCR product was mixed with 10 μl loading buffer and 10 μl loaded carefully into the slot of submerged gel, also a 100 bp DNA marker (Gibco, BRL) was loaded along as a size detector. The gel was visualized under UV light by ethidium bromide fluorescence [22]. The expected HCV-RNA band was at the level of 150 bp range, and ApoE at a 227 bp. The restriction enzyme cuts the amplified region as following: Apo-Eε3 gives 91 and 48 bp bands length while ApoE-ε2 gives 91 and 81 bp bands length and Apo-Eε4 gives 72 and 48 bp bands length (Figure 1).
Figure 1: Shows the restriction map of apoE by using CfoI restriction enzyme [22].
Concentration of apolipoprotein: All patients included in our study were tested for anti-ApoE antibodies. Apolipoprotein E specific antibody has been procoated onto 96-well plates and blocked. All samples are added to the wells and subsequently an Apolipoprotein E specific biotinylated detection antibody is added and then followed by washing with wash buffer. TMB is then used to visualize streptavidinperoxidase enzymatic reaction. TMB is stimulated by Streptavidinperoxidase to produce a blue color product that changes into yellow after adding acidic stop solution. The concentration of yellow coloration is directly proportional to the amount of Apolipoprotein E captured in plate.
HCV genotyping procedure
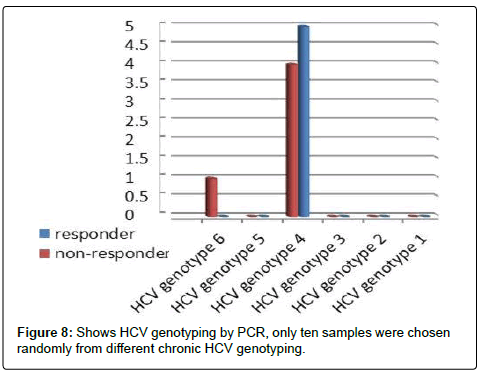
Genotyping was performed on the 10 random selected patients (5 from each respond and non-respond patients to sovaldi treatment). HCV-RNA was extracted from stored serum samples using a QIAGEN kit (QIAamp MinElute Virus Spin Extraction kit, Germany). INNO LIPA HCV (Ampliquality HCV-TS; AB-Analitica, Italy) assay based on reverse dotblot principle was used for HCV genotyping. The probes in this assay allow the identification of the following types: 1, 1a, 1b, 1a/b, 1c, 1d, 2, 2b, 2c, 3, 4, 5a, 6a, 7. Results were evaluated according to the schedule in kit insert. These results were performed at Al-Borg laboratory, Cairo, Egypt.
Statistical analysis
Data were coded and entered using the statistical package SPSS version 21. Data was summarizеd using mean, standard deviation and median, minimum and maximum for quantitative variables and frequencies (number of cases) and relative frequencies (percentages) for categorical variables. Comparison of quantitative variables was done using the nonparametric Kruskal-Wallis when comparing more than 2 groups and using the nonparametric Mann–Whitney U test when comparing 2 groups. Chi square or Fisher’s exact test was used for comparison between groups; as appropriate. Odds ratios and their 95% confident intervals were calculated. A p value ≤ 0.05 was considered statistically significant.
Results
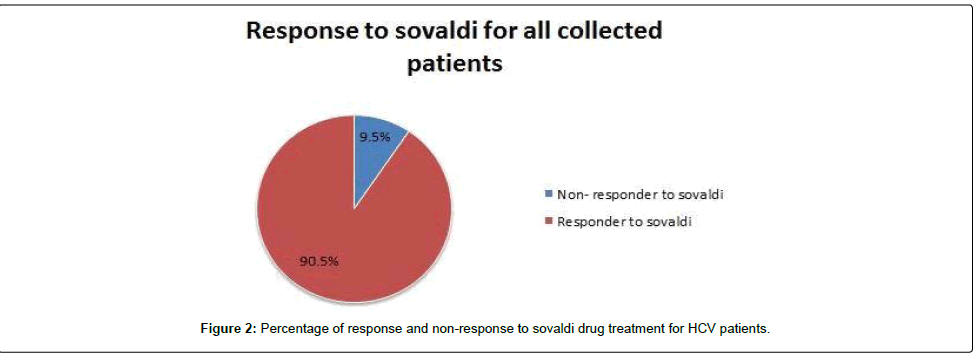
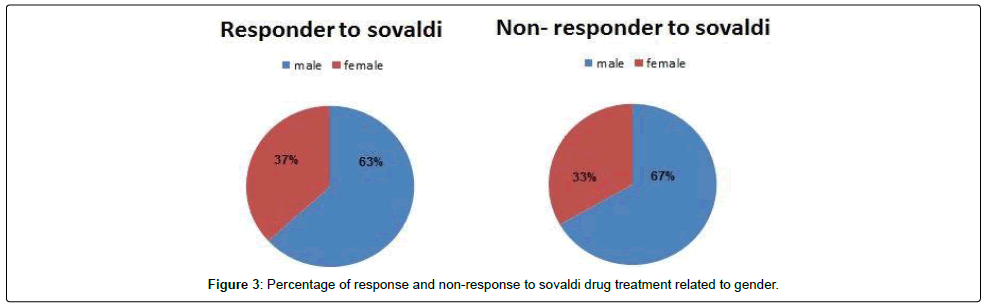
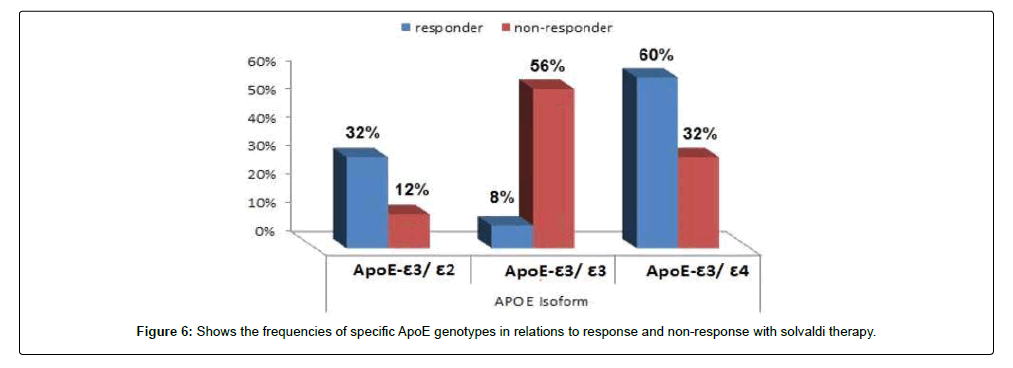
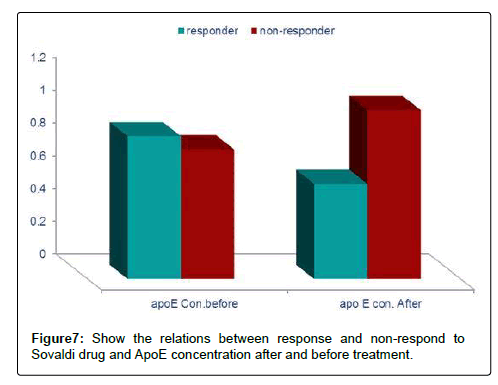
We tested 285 patients carrying anti-HCV antibodies using ELIZA Technique and confirmed HCV RNA by the reverse transcriptase polymerase chain reaction (RT-PCR). All patients were treated with sovaldi. (Table 1 and Figure 2). Our results showed that, among 285 patients were infected with Hepatitis C virus and treated with sovaldi drug 258/285 (90.5%) responded to sovaldi treatment, but only 27/285 (9.5%) did not respond to treatment (Table 2). Our results showed that for patients who responded to treatment with sovaldi drug, we found that there were 163 (63%) males and 95 (37%) females, for patients who did not respond to treatment, we found that there were 18 (67%) males and 9 (33%) females (Table 3 and Figure 3). All patients aged between 30 to 55 years, where 13 (26%) cases of females and 37 (74%) cases of males (Table 4). Out 285 patients were infected with hepatitis C virus and treated with sovaldi drug, we selected 50 patients (25 responded to sovaldi treatment and 25 no-responded to treatment (Table 5) for other investigation. Our study showed that, in the case of response to the treatment of the Sovaldi drug, there are three genotypes of apoE ε3/ε2, ε3/ε3 and ε3/ε4 were detected. ApoE genotype ε3/ε2 were recorded in 8 (32%) cases, while apoE genotype ε3/ε3 where recoded in 2 (8%) cases and apoE genotype ε3/ε4 were recorded in 15 (60%) cases. For patients who did not responded to sovaldi drug, we founded 3 (12%) cases of apoE ε3/ε2, 14 (56%) cases of apoE ε3/ε3 and 8 (32%) cases of apoE ε3/ε4. Our results showed that the concentration of apoE gene increased in the case of hepatitis C infection and also in the case of non-response to treatment, while the concentration of this gene decreased in the case of response to treatment with the sovaldi drug (Tables 4-8 and Figures 5-8). Only ten samples (five from each responded and non-responded to sovaldi treatment) samples were chosen randomly from stored samples of patients with different chronic HCV infection before treatment for detection of HCV genotypes, 9/10 (90%) were results as genotype 4 (5 in responder and 4 in non-responder patients), while 1/10 (10%) were recorded as genotype 6 in non-responder patient (Figure 8 and Table 9).
| Total | HCV-Ab. | HCV RNA | ||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| 285 (100%) | 0 | 285 (100%) | 0 | |
Table 1: Serological and molecular detection of HCV infection before treatment with sovaldi drug.
| No. of patients | Response to Sovaldi therapy | PCR results HCV RNA |
|---|---|---|
| 258 (90.5%) | Non-Responded | Positive |
| 27 (9.5%) | Responded | Negative |
Table 2: Molecular detection of HCV RNA and respond sovaldi after treatment.
| Sex | Total No. | Response to sovaldi drug | |
|---|---|---|---|
| Response | Non-response | ||
| Male | 181 | 163 (63%) | 18 (67%) |
| Female | 104 | 95 (37%) | 9 (33%) |
| Total | 285 | 258 (90.5%) | 27 (9.5%) |
Table 3: Estimation of response and non-response to sovaldi drug treatment related to gender.
Figure 4: Show the 227bp amplification products of ApoE gene, 227bp product of the ApoE gene was amplified using a pair of sequence specific primers in a standard PCR reaction. The PCR product was spotted by electrophoresis in agarose gel with ethidium bromide staining. The presence of bands indicates the presence of target gene.
| Parameters | Response to Sovaldi | Significant | |||||
|---|---|---|---|---|---|---|---|
| Responder | Non-responder | Test | p value | ||||
| Count | % | Count | Column | ||||
| Sex | Male | 17 | 46% | 20 | 54% | 0.936 | 0.333 |
| Female | 8 | 61.50% | 5 | 38.50% | |||
| HCV RNA | Positive | 0 | 0.0% | 25 | 100.0% | 50 | <0.001 |
| Negative | 25 | 100.0% | 0 | 0.0% | |||
Table 4: Distribution of HCV markers after treatment with solvaldi therapy related to gender for selected random samples.
| ApoE Isoform | Sex | Significant | ||||
|---|---|---|---|---|---|---|
| Male | Female | Test | P value | |||
| Count | % | Count | % | 0.49 | 1 | |
| ε3/ ε2 | 6 | 35.30% | 2 | 25.00% | ||
| ε3/ ε3 | 1 | 5.90% | 1 | 12.50% | ||
| ε3/ ε4 | 10 | 58.80% | 5 | 62.50% | ||
| Total | 17 | 100% | 8 | 100% | ||
Table 6: Distribution of apoE genotypes related to gender in respond patient to sovaldi treatment.
| ApoE isoform | Sex | Significant | ||||
|---|---|---|---|---|---|---|
| Male | Female | Test | P value | |||
| Count | % | Count | % | 0.632 | 0.808 | |
| e3/e2 | 2 | 10.00% | 1 | 20.00% | ||
| e3/e3 | 11 | 55.00% | 3 | 60.00% | ||
| E3/e4 | 7 | 35.00% | 1 | 20.00% | ||
| Total | 20 | 100% | 5 | 100% | ||
Table 7: Distribution of apoE genotypes related to gender in non- respond patient to Sovaldi treatment.
| ApoE concentration |
Response to Sovaldi | test | P value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Responder | Non –responder | |||||||||||
| Mean | SD | Median | Minimum | Maximum | Mean | SD | Median | Minimum | Maximum | |||
| ApoE Con.before treatment |
0.87 | 0.58 | 1.01 | 0.07 | 1.99 | 0.79 | 0.60 | 0.86 | 0.06 | 2.01 | -0.583 | 0.56 |
| ApoE con. After treatment |
0.58 | 0.36 | 0.66 | 0.07 | 1.01 | 1.03 | 0.66 | 1.00 | 0.09 | 2.83 | - 2.718 |
0.007 |
Table 8: Show concentration of apoE related to response and non-response to Sovaldi drug after and before treatment.
| HCV genotyping by PCR | Response to Sovaldi | |
|---|---|---|
| Responder | Non-responder | |
| HCV genotype 1 | 0 | 0 |
| HCV genotype 2 | 0 | 0 |
| HCV genotype 3 | 0 | 0 |
| HCV genotype 4 | 5 | 4 |
| HCV genotype 5 | 0 | 0 |
| HCV genotype 6 | 0 | 1 |
Table 9: Shows HCV genotyping by PCR, only ten samples were chosen randomly from different chronicHCV genotyping.
Discussion
ApoE plays a key role in carrying of cholesterol and other lipids throughout different cells of various tissues. ApoE, a key component of several lipoprotein classes, is created by hepatocytes, but also by macrophages [23], and glia cells [24]. Polymorphism at the apoE gene locus results in the synthesis of 3 protein isoforms called ε2, ε3, and ε4, differing by a single amino acid substitution but displaying biologic properties not entirely similar [25]. Infectious low-density HCV virions were rich in ApoE alleles; the levels of ApoE and HCV RNA in infectious fractions were positively linked. A small interfering RNA-mediated (siRNA) of ApoE expression reduced ApoE secretion and caused an apparent dose-dependent suppression of HCV virions production in the cells. However, the amount of HCV RNA containing particles was not significantly affected by an ApoE over expression indicating that the level of apoE expression is not a limiting factor for HCV replication [26]. In particular, carriage of the ε4 allele is known to be associated with increased serum levels of lowdensity lipoproteins (LDLs) [27]. Previous studies have shown that HCV particles circulate in blood bound to LDL and very-low-density lipoproteins [18]. Present our study results showed that, among 285 patients were infected with Hepatitis C virus and treatment with sovaldi drug 258/285 (90.5%) were responded to treatment of sovaldi drug, but only 27/285 (9.5%) did not respond to treatment. Out 285 patients were 163 (63%) males and 95 (37%) females, while in the case of non-response to treatment of Sovaldi drug found 18 (67%) male and 9 (33%) female. This finding was notified by several others [28-30].
All patients aged between 30 to 55 years, where 13 (26%) cases of females and 37 (74%) cases of males. Our study showed that in the case of response to the treatment of the Sovaldi drug, there are three genotypes of apoE ε3/ε2, ε3/ε3 and ε3/ε4 where there are 8 (32%) cases of ε3/ε2, 2 (8%) cases of ε3/ε3 and 15 (60%) cases of ε3/ ε4. While, for patients who did not responded to sovaldi drug, we founded 3 (12%) cases of apoE ε3/ε2, 14 (56%) cases of apoE ε3/ ε3 and 8 (32%) cases of apoE ε3/ε4. Our results showed that the concentration of apoE gene increased in the case of hepatitis C infection and also in the case of non-response to treatment, while the concentration of this gene decreased in the case of response to treatment with the sovaldi drug. The ways in which ApoE interacts with HCV might need to be rethought, especially considering its role in the penetration of virus in extra-hepatic sites. In evaluating the factors associated with the response to INF-Rib combination therapy in HCV infection, prominent roles for different age and sex were suggested by some authors [22,31,32]. In our study we found that apoE-ε4 plays an important role in predicting antiviral drugs for chronic hepatitis C virus patients. Our results showed that a viral response in 15 (60%) of 25 responder patients for Sovaldi therapy had apoE-ɛ4. In agreement with previous investigation of the influence of apoE polymorphism and outcome of HCV infection suggested that the ApoE-4 allele protected against sever liver disease [33]. In contrary, other results have suggested that carriage of the ApoE-ε4 allele is not associated with better viral response rate after treatment of HCV infection [34]. Our study was in contrast with other results suggested that apoE-ε2 allele was associated with an increased likelihood of viral clearance and interestingly, there were no HCV antibody positive patients with apoE-ε2 genotype, although this is relatively rare in healthy control [35]. Another study suggested that heterozygosity for the apoE-ε4 allele might be associated with better histological outcome in current HCV infection in the liver transplantation setting [36], but this situation is complicated by possible differences in donor/recipient ApoE alleles. It is also of interest that chronic HCV is associated with cognitive dysfunction [35], and there is study demonstrate an association between apoE-ε4 and neuropsychiatric symptoms during interferon alpha treatment for chronic HCV [37]. Our study demonstrated that apoE-ε3 allele was the highest detectable allele among patients, 14 out of 50 patients was had apoE-ε3. Among whom 14 (56%) of 25 had apoE-ε3 are not responded to Sovaldi therapy, these results reported here may indicate an association between the apoE-ɛ3 allele and persistence of hepatitis C virus. This would fit a model whereby apoE-ɛ3 binds to receptors with high affinity and is associated with normal serum cholesterol and triglyceride levels, this finding were reported by others [38]. In the present study, these two factors are not significantly related to apoE genetic polymorphisms and patients antiviral treatment. Other results showed that HCV linked to apoE-ε4 helps the virus avoid neutralization by antibodies against HCV isolated from chronically infected patients. This method of immune evasion poses a challenge for the development of HCV vaccines [39]. Our results founded that from ten samples which selected randomly from stored samples with chronic HCV, 9/10 (90%) samples were recorded as HCV genotyping 4 (5 in responder and 4 in non-responder patients), only one sample was recorded as HCV genotyping 6 (non-responder patients). Several oral direct-acting antiviral (DAA) combination regimens for the treatment of genotype 4 HCV (e.g., sofosbuvir plus ledipasvir, sofosbuvir plus simeprevir, or paritaprevir/ritonavir plus ombitasvir with our without ribavirin) were evaluated in many studies, which reported high sustained virologic response (SVR) rates with few side effects [39,40]. Sofosbuvir plus daclatasvir combination therapy was extensively investigated in HCV genotypes 1, 2, and 3, while published data regarding its real-life application in the treatment of genotype 4, particularly in Egypt, is lacking [41].
Conclusion
These findings may yield insights into the pathogenesis of HCV disease. Knowledge of the apoE Concentration and genotype of an individual with chronic HCV infection may influence clinical management in situations in which resources are not sufficient to allow treatment of all patients who might otherwise require it. It is important to determine the genotype of apolipoprotein E for all patients with hepatitis C virus to determine the degree of probability of response to systems used in the treatment of chronic hepatitis C virus.
References
- Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST (2013) Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57: 1333-1342.
- Webster DP, Klenerman P, Dusheiko GM (2015) Hepatitis C. Lancet 385: 1124-1135.
- Barth H, Liang TJ, Baumert TF (2006) Hepatitis C virus entry: molecular biology and clinical implications. Hepatology 44: 527-535.
- Huang H, Sun F, Owen DM, Li W, Chen Y, et al. (2007) Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA 104: 5848-5853.
- Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF (2011) Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol 54: 566-576.
- Kim DS, Burt AA, Ranchalis JE, Jarvik ER, Rosenthal EA, et al. (2013) Novel gene-by-environment interactions: APOB and NPC1L1 variants affect the relationship between dietary and total plasma cholesterol. J Lipid Res 54: 1512-20.
- Aizawa Y, Seki N, Nagano T, Abe H (2015) Chronic hepatitis C virus infection and lipoprotein metabolism. World J Gastroenterol 21: 10299-313.
- Lin WR, Graham J, MacGowan SM, Wilcock GK, Itzhaki RF (1998) Alzheimer’s disease, herpes virus in brain, apolipoprotein E4 and herpes labialis. Alzheimers Rep 1: 173-178.
- Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, et al. (2002) Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol 155: 487-95.
- National Center for Biotechnology Information, Database Single Nucleotide Polymorphism.
- Radmanesh F, Devan W, Anderson C (2014) Accuracy of imputation to infer unobserved APOE epsilon alleles in genome-wide genotyping data. Eur J Hum Genet 22: 1239-42.
- Huebbe P, Lodge JK, Rimbach G (2010) Implications of apolipoprotein E genotype on inflammation and vitamin E status. Mol Nutr Food Res 54: 623-30.
- Mahley RW, Rall SC (2000) Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet 1: 507-537.
- Corder EH, Robertson K, Lannfelt L (1998) HIV-infected subjects with the E4 allele for APOE have excess dementia and peripheral neuropathy. Nat Med 4: 1182-1184.
- Wozniak MA, Itzhaki RF, Faragher EB (2002) Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology 36: 456-463.
- Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, et al. (2008) Cellular determinants of hepatitis C virus assembly, maturation, degradation and secretion. J Virol 82: 2120-2129.
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, et al. (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089-1097.
- Thomssen R, Bonk S, Propfe C, Heermann KH, Kochel HG, et al. (1992) Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol (Berl) 181: 293-300.
- Ye J (2007) Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis c virus.PloS Pathog 3: 108.
- Burlone ME, Budkowska A (2009) Hepatitis C virus cell entry: role of lipoproteins and cellular receptors.J General Virol 90: 1055-1070.
- Nickerson DA, Taylor SL, Fullerton SM, Weiss KM, Clark AG, et al. (2000) Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res 10: 1532-1545.
- De Wali M, Harrison RF, Gow PJ (2002) Advancing donor liver age and rapid fibrosis progression following transplantation for hepatitis C. Gut 51: 248-252.
- Werb Z, Chin JR, Takemura R (1986) The cell and molecular biology of apolipoprotein E synthesis by macrophages. Ciba Found Symp 118: 155-171.
- Pitas RE, Boyles JK, Lee SH (1987) Astrocytes synthesize apolipoprotein E and metabolize polipoprotein E–containing lipoproteins. Biochim Biophys Acta 917: 148-161.
- Linton MF, Gish R, Hubl ST (1991) Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J Clin Invest 88: 270-281.
- Chang KS, Jiang J, Cai Z, Luo G (2007) Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol 81: 13783-13793.
- Sing CF, Davignon J (1985) Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet 37: 268-285.
- Lavezzo B,Franchello A, Smedile A (2002) Treatment of recurrent hepatitis C in liver transplants: efficacy of a six versus a twelve month course of interferon alfa 2b with ribavirin. J Hepatol 37: 247-252.
- De Vera ME, Smallwood GA, Rosado K (2001) Interferon alpha and ribavirin for the treatment of recurrent hepatitis C after liver transplantation. Transplantation 71: 678-686.
- Machicao VI, Bonatti H, Krishna M (2004) Donor age affects fibrosis progression and graft survival after liver transplantation for hepatitis C. Transplantation 77: 84-92.
- El-Adly AM, Wardany AA (2017) Seroprevalence of Hepatitis C Virus among Population in Luxor Governorate, Egypt. J Hum Virol Retrovirol 5: 00144
- Wozniak MA, Itzhaki RF, Faragher EB (2003) Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology 36: 456-463.
- Mueller T, Gessner R, Sarrazin C, Graf C, Halangk J (2003) Apolipoprotein E4 allele is associated with poor treatment response in hepatitis C virus (HCV) genotype 1. Hepatology 38: 1592.
- Price DA, Bassendine MF, Norris SM, Golding C, Toms GL (2006) Apolipoprotein epsilon3 allele isassociated with persistent hepatitis C virus infection. Gut 55: 715-718.
- Fabris C, Vandelli C, Toniutto P, Minisini R, Colletta C (2011) Apolipoprotein E genotypes modulate fibrosis progression in patients with chronic hepatitis C and persistently normal transaminases. J Gastroenterol Hepatol 26: 328-33.
- Gochee PA, Powell EE, Purdie DM, Pandeya N, Kelemen L (2004) Association between apolipoprotein E epsilon4 and neuropsychiatric symptoms during interferon alpha treatment for chronic hepatitis C. Psychosomatics 45: 49-57.
- Catherine F, Daniel JF, Emilie C, JiYoung L (2015) Apolipoprotein E Mediates Evasion From Hepatitis C Virus Neutralizing Antibodies. Gastroenterology 21: 1-10.
- Kohli A, Kapoor R, Sims Z (2015) Ledipasvir and sofosbuvir for hepatitis C genotype 4: a proof-of-concept, single-centre, open-label phase 2a cohort study. Lancet Infect Dis 15: 1049-1054.
- Asselah T, Hassanein TI, Qaqish RB (2015) Efficacy and safety of ombitasvir/paritaprevir/ritonavir co-administered with ribavirin in adults with genotype 4 chronic hepatitis C infection and cirrhosis (Agate-I). Hepatology 62: 119.
- Asselah T (2014) Daclatasvir plus sofosbuvir for HCV infection: an oral combination therapy with high antiviral efficacy. J Hepatol 61: 435-438.
- Fontaine H, Hezode C, Zoulim F (2015) Efficacy of the oral sofosbuvir-based combinations in HCV genotype 4-monoinfected patients from the French observational cohort ANRS CO22 Hepather. J Hepatol 62: 278.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi