Review Article, J Sleep Disor Treat Care Vol: 12 Issue: 1
Sleep Apnea Events Detection Using Deep Learning Techniques
Mahmood Abed1* and Turgay Ibrikci2
1Department of Electrical and Electronics Engineering, Gaziantep University, Gaziantep, Turkey
2Department of Software Engineering, Adana Science and Technology University, Adana, Turkey
*Corresponding Author: Mahmood Abed
Department of Electrical and Electronics
Engineering, Gaziantep University, Gaziantep, Turkey
Tel: +909647823163763
E-mail: mahmood.abed882@gmail.com
Received date: 02 January, 2023, Manuscript No. JSDTC-23-75292;
Editor assigned date: 05 January, 2023, PreQC No. JSDTC-23-75292 (PQ);
Reviewed date: 27 January, 2023, QC No. JSDTC-23-75292;
Revised date: 03 February 2023, Manuscript No. JSDTC-23-75292 (R);
Published date: 10 February, 2023, DOI: 10.4172/2325-9639.23.12.101
Citation: Abed M, Ibrikci T (2023) Sleep Apnea Events Detection Using Deep Learning Techniques. J Sleep Disor Treat Care 12:1.
Abstract
This research underlines an automated approach for detecting sleep apnea events from sleep studies. The Polysomnogram test is the gold standard for diagnosing sleep apnea. Unfortunately, it is expensive, time-consuming, and uncomfortable for patients. We selected signals that can be simply obtained by using a portable fingertip pulse oximeter and hexoskin smart shirt. Hence, the cost of polysomnography will be reduced by utilizing less equipment and sufficient at the same time. Therefore, the scientific value of this research is to simplify the used ways by other sleep experts in this field. Two sleep apnea databases were used to train and test four deep learning models. Three physiological signals were combined to form one window of 60 seconds in size. Deep learning approaches were proved to be sufficient in detecting apnea events depending on data quality and the neural network architecture. The hybrid model outperformed other models with 97% and 92% of accuracy.
Keywords: Sleep apnea; Polysomnography; Deep learning
Introduction
Sleep apnea is a respiratory-related disease where breathing pauses and repeatedly starts during sleep. It occurs when either the airway collapses or the brain cannot successfully send the signal to the breathing muscles. There are two main cases for sleep apnea; Obstructive Sleep Apnea (OSA) and Central Sleep Apnea (CSA), which in no air comes in or out of the lung for few seconds to minutes and can, happen about 30 times or more per hour [1]. Hypopnea is a partial blockage of the airway with at least a 30% decrease in airflow enduring at least 10 seconds and with 3% oxygen desaturation [2]. Neglecting the treatment of sleep apnea leads to severe diseases, such as high blood pressure and heart attack [3].
Polysomnography (PSG) is used to diagnose the patient who spends an entire night or more in a sleep laboratory. PSG is often ambulatory, so patient can sleep at home. However, sleeping in the lab may occur uncomfortable because of the connection of the electrodes with different positions of the body [4]. Physiological signals are divided into two categories: Simple signals measured by sensors that are integrated into smart wearable devices and complex signals obtained by professional tools while transferred to user devices. The simple signals are the heartbeat and snoring, whereas the complex ones include Electrocardiography (ECG), Electroencephalography (EEG), Electromyography (EMG), Electrooculography (EOG), Oxygen Saturation (SpO2), Nasal airflow, and blood pressure. sleep experts’ diagnose this disease by monitoring and analyzing these signals during the total time of sleep. The current treatment is the titration of continuous positive Airway Pressure (CPAP), Bilevel Positive Airway Pressure (BiPAP) and Adaptive Servo Ventilation (ASV). They control the airway and keep it open continuously [5]. This study aims at determining whether deep learning can detect sleep apnea events from any PSG. This will add value to future researches.
Literature review
Many researchers conducted studies on sleep apnea events detection differently. Some of them used feature engineering with traditional machine learning methods whereas others adapted deep learning techniques [6]. Demonstrated Long Short-Term Memory (LSTM) with a single ECG signal. The apnea diagnosis was carried out on the characteristics derived from the Heart Rate Variability (HRV) tests. Long Short-Term Memory (LSTM) was chosen to report the time based dependency in HRV data, as one of the critical aids of LSTM is the capability to use the prior situation Suggested using LSTM to detect the OSA severity by using one feature Instantaneous Heart Rate (IHR) alone [7]. Then, by adding an extra feature, which is the SpO2. Many physicians applied this technique to their patients [8-10]. Reported Convolutional Neural Network (CNN) architecture to detect apnea with different trials. First, they used the nasal airway signal to equate CNN to Support Vector Machine (SVM). Later, the 2D spectrogram images of the nasal airflow signal with the raw 1D nasal airway signal were used to apply a different strategy. Their results adapted using three separate signals; nasal airflow, abdominal and thoracic. Urtnasan used a single ECG signal with CNN, LSTM, and Gated Recurrent Unit (GRU) techniques. They recorded high performance [11,12]. However, they could have proved that by using multisignal generalized the study by training the RCNN model on MGH data and testing it on SHHS data, though this approach lacked accuracy. Li proposed a novel approach for improving accuracy by integrating the Hidden Markov Model (HMM) with Deep Neural Network (DNN), while adopting the fusion decision algorithm to boost overall performance [13]. Nesaragi proposed an LSTM model consisting of two stages while neglecting the SpO2 signal [14]. They discovered the strong point of using instantaneous frequency and spectral entropy features for the detection of arousals. Islam proposed another way of detecting sleep apnea using 3D scans while using predefined models in transfer learning to beat the limitation of the small data set. Their results illustrated the connection between facial morphology and OSA. Wang developed an improved LeNet-5 convolutional neural network with ECG segments for sleep apnea detection [15]. The use of transfer learning models has proved useful and promised. Mahmud referred to low performance caused by the data’s lack. They used EEG signals composed of only seven different apnea patients [16].
Material and Methods
This part illustrated the utilized sleep apnea databases and the used deep learning algorithms with the performance metrics of this study.
Sleep apnea databases
Two sleep apnea databases from PhysioNet are used for this study [17]. These datasets are "You Snooze You Win Database" was a target at the PhysioNet/Computing in cardiology challenge 2018 is provided by the Massachusetts Genearl Hospital (MGH) Computational Clinical Neurophysiology Laboratory (CCNL) and the Clinical Data Animation Center (CDAC) [18]. It includes 1,985 patients for the detection of sleep disorders obtained in an MGH sleep laboratory. They split the database into 994 folders as a training set and 989 folders as a test set. Certified sleep technologists at the MGH labeled the database according to the existence of arousals. These derived arousals were either classified as: Respiratory Effort-Related Arousals (RERA), spontaneous arousals, hypoventilation, bruxism, hypopneas, apneas (central, obstructive, and mixed), vocalizations, snores, periodic leg movements, breathing cheyne stokes or partial airway obstructions [19]. The database includes two directories (training and test). Each directory contains one sub-folder per patient. Every subfolder includes signal, header, and arousal files. Test sets are unlabeled. Therefore, we were unable to use them in the testing. Records in the database were taken from different persons in both sex and age.
This challenge’s database “Apnea-ECG Database (Challenge 2000)” includes 70 records, divided into two equal parts, 35 records as a training set (a01-a20, b01-b05, and c01-c10), and 35 records as a test set (x01-x35) [20]. Only the learning sets were annotated. Each record includes signals, headers, and other data. Besides, eight records (a01, b01, c01, a02, c02, a03, c03, and a04) are followed by four additional signals (CHEST, ABD, Nasal Airflow, and SpO2). Three of them were used for further testing [21]. Figure 1 shows the approach of sleep apnea events detection.
Deep learning techniques
Long Short-Term Memory (LSTM): Hochreiter and Schmidhuber initially suggested Long Short-Term Memory (LSTM) expression in 1997 [22]. LSTM is a kind of Recurrent Neural Network (RNNs), and it became very popular in recent years due to its significant performance and in solving the vanishing gradient problem. LSTM overcomes that by imposing fixed error flow. It clearly learns when to save the information and when to retrieve it using gradient descent. LSTM has distinct elements in the recurrent hidden layer, called memory blocks. These blocks provide memory cells with self-connections to preserve the network’s time based state as well as different multiplicative units called gates to alter information flow [23].
Each memory block within the interior architecture consists of three types of gates, as shown in Figure 2, which are specifically:
Input gate: It dominates the quantities that go into the cell of the new value.
Output gate:It considers the input at time t, the prior hidden state, and the present value of the cell.
Forget gate: It dominates the ex-cell value quantities that go into the present cell value.
The memory cell in an LSTM network works as a single unit within the hidden layer of traditional networks. The formulas are given in the following equations:

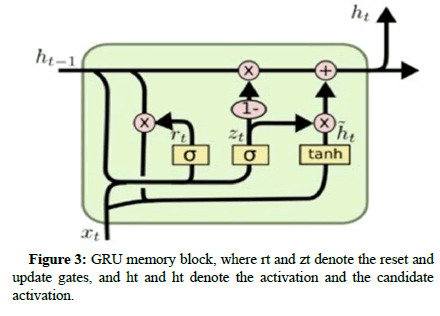
Gated Recurrent Unit (GRU): Many similar concepts, but it has a much smaller set of parameters so that it can be trained more quickly at a sustained hidden layer size. Some researches illustrated that the accuracy between LSTM and GRU is comparable and even better with the GRU in some cases. GRU includes the same internal structure of LSTM, but the memory cell consists of two gates rather than three, as shown in Figure 3, which are namely:
Update gate: It also compares how much of the hidden value of the previous candidate and how much of the hidden value of the current candidate combines to get the new hidden value.
Reset gate: It controls how much of the previous hidden state is considered when the new candidate hidden value is created. In other words, it can “reset” the hidden value.

Residual Network (ResNet): Residual Network (ResNet) is a pretrained deep neural network model used in many tasks like prediction and feature extraction, or being fine-tune to a specific case. Transfer learning is wisely using the knowledge acquired earlier from different missions or issues to mitigate new problems quicker [24]. ResNet50 model was used as the pre-trained model for sleep apnea events detection by eliminating the predicting layer and substituted it with our binary predicting layer. Weights of the first few layers were untouched or updated during the training because they save general information like curves and edges. Instead, we made the network to emphasis on learning specific features in the subsequent layers [25].
Hybrid model: It consists of the ResNet50 architecture with two RNN layers. One GRU layer comes after the input layer, and one bidirectional LSTM layer comes before the output layer.
Performance assessments: The test set includes samples that have never been seen before by the algorithm. Therefore, if the model performs well in predicting, it can be assumed that it is generalizing well. We used the following metrics for assessing the classification model:

Results
Some standard python packages were used to access physionet. They were also used to display and prepare signals for apnea events detection. We used the WFDB API package for remote access instead of downloading the training set for data preprocessing [26]. We obtained 13 physiological signals in each PSG. Signals were measured in microvolts, excluding oxygen saturation (SpO2), which was measured as a percentage. We selected three signals (ABD, CHEST, and SpO2) and dropped the rest from our estimations. ABD and CHEST refer to abdominal and thoracic belts, respectively. An apnea events dictionary was created, containing only apnea labels [27]. All labels for apnea/stages were pulled from PhysioNet and combined with those three signals. They were loaded into a data frame, and then the apnea events dictionary that we created was mapped with the same data frame. Hence, other unused labels (sleep stages) in that dictionary were shown as missing values. Therefore, they were replaced by zero [28]. Labels are finally encoded into the one-hot array.
We used google collaborator as a free cloud service based on jupyter notebooks for implementing this task. Colab gives 12 GB of RAM and increases it up to 25 GB after runtime if required [29]. We downloaded the training set of the YSYW database directly, but in multistages to avoid RAM crashing. The figures illustrate the CHEST signal annotated by an obstructive apnea and the raw targeted signals before preprocessing (Figures 4-8).
Signals where SpO2 is below 50% were removed because there is a possibility that the connection with devices would be lost [30]. The sampling frequency was resampled to 100 Hz because 100 samples per second are enough to use. The imbalanced problem between apnea and normal classes was fixed by under sampling. Finally, we reshaped signals into intervals with a fixed length [31]. We applied the minimum-maximum method to scale data between 0 and 1 for normalization according to equation 15. Figure 6 shows the final shape of signals before using them for the task of apnea events detection (Table 1) [32].
| Databases | Models | True positive | True negative | False positive | False negative |
|---|---|---|---|---|---|
| YSYW | LSTM | 8507 | 25473 | 2715 | 2514 |
| Apnea-ECG | LSTM | 1402 | 1065 | 209 | 173 |
| YSYW | GRU | 9507 | 26622 | 1715 | 1365 |
| Apnea-ECG | GRU | 1195 | 1410 | 155 | 89 |
| YSYW | ResNet50 | 9067 | 23906 | 2980 | 3247 |
| Apnea-ECG | ResNet50 | 1219 | 1183 | 257 | 190 |
| YSYW | Hybrid | 10433 | 27433 | 1004 | 329 |
| Apnea-ECG | Hybrid | 1440 | 1193 | 174 | 42 |
Table 1: Confusion matrices parameters of our models applied to the YSYW and Apnea-ECG databases. Note: 0: Normal and 1: Apnea.
Several techniques were chosen for comparison according to their performances. LSTM and GRU models included four LSTM and GRU layers, respectively. Each has 30, 60, 120, or 200 memory cells followed by batch normalization and dropouts to avoid over fitting. For classification, we used one fully connected layer at the output layer. ResNet50 model included 48 convolution layers, one maxpool layer, and one average pool layer. Convolution layers related to different kernel size and activation functions. An average pool layer followed by a fully connected layer containing 1000 nodes with softmax function at the end.
We extracted 39209 windows from the test set of the YSYW database, which represents the last 91 patients of the training set. Besides, we also extracted 2849 windows from the test set of the Apnea-ECG database, which represent only eight patients who have signals of ABD, CHEST, and SpO2. These windows were gathered from every patient’s data. Each window was formatted as a 1 × 6000. The mathematical meaning of this representation is that the size of the sampling window, which is 60 seconds, was multiplied by the size of the sampling frequency. The amplitude of the signals was normalized between 0 and 1. In the training phase, we divided the PSG data of each patient into short intervals and shifted these intervals for data augmentation. The best validation accuracy started to be stable beyond 700 epochs of training [33]. The batch size was taken as 32 because a big batch size would lead to poor generalization and lower test accuracy. We applied more than one deep learning model in order to compare the prediction and generalization performance with other methods. According to Tables 1 and 2 it can be concluded that the hybrid model gives the best results in both databases with an overall accuracy of 97% and 92%. GRU model comes next with an overall accuracy of 92% and 91%.
| Databases | Signals | Models | Train Subjects | Test Subjects | Accuracy (%) | Precis (%) | Rioencall (%) | F1-score (%) |
|---|---|---|---|---|---|---|---|---|
| YSYW | ABD CHEST Spo2 | LSTM | 900 | 91 | 87 | 76 | 77 | 76 |
| pnea-ECG | ABD CHEST Spo2 | LSTM | - | 8 | 87 | 87 | 89 | 88 |
| YSYW | ABD CHEST Spo2 | GRU | 900 | 91 | 92 | 85 | 87 | 86 |
| Apnea-ECG | ABD CHEST Spo2 | GRU | - | 8 | 91 | 89 | 93 | 91 |
| YSYW | ABD CHEST Spo2 | ResNet50 | 900 | 91 | 84 | 75 | 75 | 75 |
| Apnea-ECG | ABD CHEST Spo2 | ResNet50 | - | 8 | 84 | 83 | 87 | 85 |
| YSYW | ABD CHEST Spo2 | Hybrid | 900 | 91 | 97 | 91 | 97 | 94 |
| Apnea-ECG | ABD CHEST Spo2 | Hybrid | - | 8 | 92 | 89 | 97 | 93 |
Table 2: Results of comparison between different deep learning models in sleep apnea events detection.
Discussion
Considering the results that we obtained in this study; we could say that deep learning has given high performance in this task. The four deep learning techniques, including LSTM, GRU, ResNet-50, and the hybrid model, were structured and evaluated according to several metrics. In practice, we found that models obtained by using only one signal from the PSG study cannot be generalized for further use because the symptoms are likely to vary based on the physical variations in patients. Consequently, it was discovered that using more than one signal gives the chance to catch a higher number of abnormal events. We used the per window method in generating signals and detection of apnea events. Some researchers used the perrecord method. They computed the Apnea Hypopnea Index (AHI) for some windows in order to obtain a record that is classified as having normal, mild, moderate, or severe apnea. Nesaragi also used the YSYW database in a different way of us [25]. First, they focused on arousal and non-arousal events. EEG signals must be used to detect arousal events. Hypopnea and arousal events can only be distinguished with EEG signals. Second, the evaluation was not based on precision or accuracy. They opted for AUROC instead and obtained low performance. They did not discuss the internal architecture of the LSTM model [34]. They trained two layers of Quadratic Discriminants (QD), which were connected to several LSTMs. Then, the output of the trained QD layers was averaged to get the final prediction. The YSYW database was not used previously for apnea events detection, which reflects our novelty and uniqueness of the results. Also, none of previous researches make use of the same group of signals that are used. However, most previous researchers preferred to use the Apnea- ECG database. Another new point of this study, learning parameters of trained models is transferred directly instead of training from scratch [35]. Finally, the window’s size, which represents the input data, is a vital feature for increasing or decreasing the performance of the system. A window of size 30 seconds might give better results because more than one apnea might occur during one minute. That is what Pomprapa emphasized in their paper [36].
Conclusion
Sleep apnea syndrome is a serious disease with complaints that cannot be relieved without treatment. The cost and time of diagnosis are exorbitant. Hence, deep learning techniques can be used in this field to provide the necessary solutions. The trained model of this study can be reused in a sleep lab or home test. The patient can collect the same data by using sensors, which can be easily obtained. On the other hand, sleep technologists can also compare their diagnosis with the predicted diagnosis to improve accuracy. The classification of sleep stages is very important for researchers. Sleep stages are obtained by analyzing EEG signals that illustrate brain activity, considering its importance in diagnosing other diseases like epilepsy. EEG can determine whether a person is asleep or awake during the sleep apnea test. Collecting labeled patient’s data is a critical problem that faced researchers in this field because one PSG needs one day to be labeled by a sleep technician.
Compliance with Ethical Standards
This type of study does not require informed consent.
Acknowledgment
The authors express thanks to the Brazilian medalist paralympic athlete (JGS) and her technical team for the availability and commitment in participating in this case study.
References
- Berry RB (2012) Rules for scoring respiratory events in sleep: Update of the 2007 AASM nanual for the scoring of sleep and associated events. J Clin Sleep Med 8: 597–619.
[Crossref] [Google Scholar] [Indexed]
- Novak D, Mucha K, Al-Ani T (2008) Long short-term memory for apnea detection based on heart rate variability. Annu Int Conf 5234–5237.
[Crossref] [Google Scholar] [Indexed]
- Pathinarupothi RK, Vinaykumar R, Rangan E, Gopalakr-ishnan E (2017) Instantaneous heart rate as a robust feature for sleep apnea severity detection using deep learning. IEEE EMBS Int Conf Biomed Heal.Informatics 293–296.
- Pathinarupothi R (2017) Single sensor techniques for sleep apnea diagnosis using deep learning. IEEE Int Conf Healthc Informatics ICHI 524–529.
[Crossref] [Google Scholar] [Indexed]
- Haidar R, Koprinska I, Jeffries B (2017) Sleep apnea event detection from nasal airflow using convolutional neural networks. JCSM 819–827.
[Crossref] [Google Scholar] [Indexed]
- Mccloskey S, Haidar R, Koprinska I, Jeffrie B, Phung D et al. (2018) Detecting hypopnea and obstructive apnea events using convolutional neural networks on wavelet spectrograms of nasal airflow. PAKDD 10937: 361–372.
- Haidar R, Mccloskey S, Koprinska I, Jeffries B (2018) Convolutional neural networks on multiple respiratory channels to detect hypopnea and obstructive apnea events. IJCNN, 1–7.
- Urtnasan E, Park JU, Lee S, Lee KJ (2017) Optimal classifier for detection of obstructive sleep apnea using a heartbeat signal. Int J fuzzy log 17: 76–81.
- Urtnasan E, Park JU, Joo EY, Lee KJ (2018) Automated detection of obstructive sleep apnea events from a single lead electrocardiogram using a convolutional neural network. J Med Syst 42: 104.
[Crossref] [Google Scholar] [Indexed]
- Urtnasan E, Park JU, Lee KJ (2020) Automatic detection of sleep-disordered breathing events using recurrent neural networks from an electrocardiogram signal. Neural Computing and Applications, 32: 4733–4742.
- Erdenebayar U, Kim YJ, Park JU, Joo EY (2019) Deep learning approaches for automatic detection of sleep apnea events from an electrocardiogram. Comput Methods Programs Biomed, 180:10500.
[Crossref] [Google Scholar] [Indexed]
- Biswal S (2018) Expert level sleeps scoring with deep neural networks. J Am Med Informatics Assoc 25(12): 1643–1650.
- Li K, Pan W, Li Y, Jiang Q, Liu G (2018) A method to detect sleep apnea based on deep neural network and hidden markov model using single lead ECG signal. Neuroma putting 294: 94–101.
- Nesaragi N, Sharma A, Patidar S, Majumder S, Tavako K (2010) Application of recurrent neural network for the prediction of target non-apneic arousal regions in physiological signals. Comput Cardiol 459: 2–5.
- Islam SM, Mahmood H, Claxton AJAA (2018) Deep learning of facial depth maps for obstruc tive sleep apnea prediction. Int Conf Mach Learn Data Eng 154–157.
- Wang T, Lu C, Shen G, Hong F, Mahmud T et al. (2019) Real time sleep apnea event detection with deep neural network. IEEE Int Conf Biomed Eng Comput Inf Technol Heal 37–41.
- Ghassemi M (2010) You snooze you win: The physioNet/computing in cardiology challenge. Comput Cardiol 20–23.
- Penzel T, Moody GB, Mark RG, Goldberger AL, Peter J (2000) Apnea-ECG database. Comput Cardiol 255–258.
- Hochreiter S, Schmidhuber J (1997) Long short-term memory. neural computation 9: 1735–1780.
[Crossref], [Google Scholar]
- Cho K (2014) Learning phrase representations using RNN encoderdecoder for statistical machine translation. EMNLP 1724–1734.
- Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Transactions on knowledge and data engineering 22:1345–1359.
- Goldberger A (2000) Physiobank, Physiotoolkit, and Physionet: components of a new research resource for complex physiologic signals. Circulation 101: 15-20.
- Pomprapa A, Sayani MS, Anwar T, Stollenwerk A, Kowalewski S et al. (2018) Apnea detection in a contactless multisensor system using deep learning algorithm. XIII RGC 86.
- Lakhan P, Ditthapron A, Banluesombatkul N, Wilaiprasitporn T (2018) Deep neural networks with weighted averaged overnight airflow features for sleep apnea-hypopnea severity classification. InTENCON 2018-2018 IEEE Region 10 Conference 0441-0445.
- Falco ID, Pietro D, Sannino G, Cioppa G, Trunfio AD (2018) Deep neural network hyper-parameter setting for classification of obstructive sleep apnea episodes. IEEE Sym-posium on Computers and Communications 1187–1192.
- Steenkiste TV, Groenendaal W, Deschrijver D (2019) Automated sleep apnea detection in raw respiratory signals using long short-term memory neural networks. IEEE J Biomed Health Inform 23: 2354–2364.
- Banluesombatkul N, Wilaiprasitporn R (2018) Deep neural networks with weighted average overnight airflow features for sleep apnea-hypopnea severity classification. TENCON 441–445.
- Cen L, Yu ZL, Kluge T, Ser W (2018) Automatic system for obstructive sleep apnea events detection using convolutional neural network. 2018 40th annual international conference of the IEEE engineering in medicine and biology society (EMBC) 3975–3978.
[Crossref] [Google Scholar] [Indexed]
- Zhang L, Fabbri D, Upender R, Kent D (2018) 0311 Automated apnea and hypopnea event detection using deep learning. Sleep, 41(1):119–120.
- Kaguara A, Nam KM, Reddy S (2014) A deep neural network classifier for diagnosing sleep apnea from ECG data on smartphones and small embedded systems. BA Computer Science.
- Chaw HT, Kamolphiwong S, Wongsritrang K (2019) Sleep apnea detection using deep learning. Tehnicvki glasnik 13:261–266.
- Alsalamah M, Amin S, Palade V (2018) Detection of obstructive sleep apnea using deep neural network. In applications of big data analytics 97–120.
- SH Choi, H Yoon, HS Kim, HB Kim, HB Kwon et al. (2018) Real time apnea hypopnea event detection during sleep by convolutional neural networks. Comput Biol Med 100: 123-131.
[Crossref] [Google Scholar] [Indexed]
- Sh MS, Mendonca F, Morgado Dias F, Ravelo Garcia A (2017) Recurrent neural network based classification of ECG signal features for obstruction of sleep apnea detection. IEEE 2:199–202.
- Dey D, Chaudhuri S, Munshi S (2018) Obstructive sleep apnea detection using convolutional neural network based deep learning framework. Biomedical engineering Letters 8: 95–100.
- Dprogrammer O, Mayoclinic O, Berry RB, Budhiraja R, Gottlieb DJ et al. (2020) Rules for scoring respiratory events in sleep: Update of the 2007 AASM manual for the scoring of sleep and associated events. J Clin Sleep Med 8: 597–619
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi 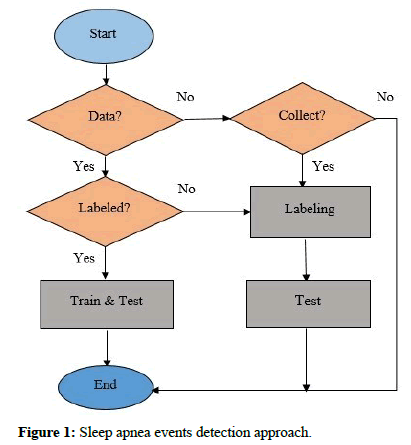


 )CHEST; (
)CHEST; ( ) resp_obstructive apnea (right).
) resp_obstructive apnea (right).
 )CHEST; (
)CHEST; ( )ABD;
(
)ABD;
( )SaO2
)SaO2
 )CHEST;
(
)CHEST;
( )ABD; (
)ABD; ( )SaO2
)SaO2