Research Article, J Immunol Tech Infect Dis Vol: 11 Issue: 4
Optimization of lateral flow assay for Canine morbillivirus detection and the application of the strip as sample substitute
Monu Karki1*, KK Rajak1, Praveen Singh2, Arfa Fayaz1, Kiran1, Mukesh Bhatt1, Vishal Rai1, Chris Einstein1, Ajay Kumar Yadav1 and RP Singh3
1Department of Biology, ICAR-Indian Veterinary Research Institute, Izatnagar, Uttar Pradesh, India
2Department of Biochemistry, ICAR-Indian Veterinary Research Institute, Izatnagar, Uttar Pradesh, India
3Department of Chemistry, ICAR, Bhubaneshwar, Odisha, India
*Corresponding Author: Monu Karki
Department of Biology,
ICAR-Indian
Veterinary Research Institute,
Izatnagar,
Uttar Pradesh,
India
Tel: +8449692979;
E-mail: karkim.91@gmail.com
Received date: 10 June, 2022, Manuscript No. JIDIT-22-66326; Editor assigned date: 13 June, 2022, PreQC No JIDIT-22-66326 (PQ); Reviewed date: 27 June, 2022, QC No. JIDIT-22-66326; Revised date: 29 August, 2022, Manuscript No. JIDIT-22-66326 (R); Published date: 07 September, 2022, DOI: 10.4172/2329-9541.1000331
Citation: Karki M, Rajak KK, Singh P, Fayaz A, Kiran, Bhatt M, Rai V, Einstein C, Yadav AK, Singh RP (2022) Optimization of Lateral Flow Assay for Canine Morbillivirus Detection and the Application of the Strip as Sample Substitute. J Immunol Tech Infect Dis 11:4.
Abstract
Canine distemper is an emerging disease, caused by the Canine morbillivirus (CDV) of the Paramyxoviridae family. The virus has evolved as a multi host pathogen as it affects many wildlife animal species. The development of specific and sensitive diagnostic tests is the need for a control program. Several diagnostic tests are available for the detection of CDV antigen and antibody. Lateral Flow Assay (LFA) is the most promising point of care diagnostic test because of its specificity, easy use and instant result. This study was designed to develop a lateral flow assay using the in-house developed monoclonal Antibody (mAb) against the Nucleocapsid protein (N) of the ‘CDV/dog/bly/Ind/2018’ isolate, which represents the circulating strains of India. The two mAbs included in the study showed high binding affinity in indirect ELISA and dot blot assay. Out of two, one mAb was selected due to its comparatively higher binding affinity in LFA format, and less non-specific binding to the biological matrix and buffer components. The fresh clinical samples collected on the spot were distinctly detected by the LFA, whereas the stored samples with a reduced titre of the virus showed inconsistent results. Moreover, the blood samples showed a clear distinction of positive and negative than the swab and tissue homogenates. The RNA extraction from the strip was successful with the some modifications in the Trizol RNA extraction method and the N and H gene fragments were amplified. Therefore, the study concludes that the LFA is suitable for CDV antigen detection in the field conditions and the strips can be used as the sample substitute for molecular study.
Keywords: Canine morbillivirus (CDV), Lateral Flow Assay (LFA), Nucleocapsid protein (N), Monoclonal Antibody (mAb), Gold Nanoparticles (GNPs), Enzyme Linked Immunosorbent Assay (ELISA)
Introduction
Canine Distemper (CD) is primarily a viral disease of dogs and can infect several wildlife animal species. The increasing host range of CDV affecting the species such as lions, leopard and red pandas has raised the concern about wildlife safety [1,2]. The control of CDV is necessary to save the endangered, vulnerable and many other wildlife species to maintain balance in the ecological system. The disease affects the respiratory, gastrointestinal and nervous systems of the infected animal and more often ends with chronic signs [3]. The chronic signs of the disease include encephalitis leading to seizures, tremors, or twitching in hind limbs, seen after around the 24th day post infection when the virus reaches and affects CNS [4].
Canine distemper virus belongs to the order Mononegavirales, family Paramyxoviridae, subfamily Orthoparamyxovirinae, and genus Morbillivirus. In 2016, ICTV renamed Canine Distemper Virus (CDV) as Canine Morbillivirus [5]. CDV is a non-segmented negative sense RNA virus of 15960 nucleotides, wrapped in a lipoprotein envelope. The genome comprises six structural proteins viz. Nucleocapsid protein (N), Phosphoprotein (P), Matrix protein (M), Fusion protein (F), Haemagglutinin protein (H), and the large protein (L) [6]. The genome of CDV is represented as 3’-N-P-M-F-H-L-5’, which also depicts the gradient of expressed proteins in infected cells in decreasing order from 3’ to 5’. Seventeen lineages of CDV are reported, with the eighteenth lineage proposed in 2019 from India [7,8]. The South America/North America–4 lineage was reported in 2020 indicating the continuous CDV emergence and its intercontinental spread [9]. China has also recently reported a new lineage from red pandas, proposed as Asia-6 [10]. Several studies indicating the genomic differences in wild type virus and vaccine strains are also reported often.
The current status of the emergence of CDV and the low efficiency of available vaccines and diagnostics are the recent concerns. New target based diagnostics and vaccines are in need to counterfeit the effects of circulating strains in respective geographical locations including India. Several diagnostic tests are available for CDV diagnosis, with varied sensitivity, specificity, required time and different levels of skills to perform the test. The problem of sample collection in wildlife and the transport related damage raises the need for specific, easy and on site diagnostic assays like LFA. In the present study, we aimed to develop a LFA for the detection of CDV using the in house developed mAbs against the N protein of CDV/dog/bly/Ind/ 2018 isolate (Unpublished). As the mAbs bind to their targets with high specificity and therefore are preferred over polyclonal antibodies for developing diagnostics [11]. The virus used in the study was isolated and characterized towards the development of vaccine candidate by our laboratory (Indian Patent application No. 202011057169). The advantage of development of a mAb based LFA will be able to detect CDV antigen across the species including wildlife animals. Also, the LFA presents an advantage over other molecular diagnostics when high throughput sampling is not required and only a single sample needs to be screened.
Materials and Methods
Up scaling and purification of monoclonal Antibodies (mAbs)
Two CDV mAb clones (A and B) against CDV (Dog)/Bly/Ind/2018 isolate developed in the laboratory were revived and up scaled by repeated hybridoma culture. Four healthy swiss albino mice (4 to 5 week old) were primed with Freund’s Incomplete Antigen (FIA) two days before inoculation of mAb clones. The animals used in the study have been approved by animal ethics committee. Hybridoma clones were centrifuged and re suspended in serum free balanced salt solution (5 × 107 cells/150 μl) and inoculated intraperitoneally [12]. The ascites fluid was collected by paracentesis when the abdominal distension was prominent in mice. The different taps of ascites fluid collected were pooled together, filtered (45 μm pore size Milipore membrane filter), and properly labeled aliquots of different volumes were made and stored at –20℃.
Ascites fluid containing mAb was purified using the standard protocol of the montage antibody purification kit. Eluted antibodies were dialyzed to remove the salts of elution buffer, then re suspended and concentrated in 0.1 mM PBS, pH 7.5 using a 30 kDa m.wt cut off Milipore Amicon filter. The reactivity of the purified and concentrated mAbs was again confirmed and the final titre was estimated using iELISA. The concentration of each antibody was estimated using the BCA™ protein assay kit.
Antigen-antibody interaction study
The bio interaction of CDV antigen and mAb in Nitrocellulose Membrane (NCM) was assessed by dot blot assay at 1 min and 5 min incubation. The 2 μl of ultra purified CDV antigen, CDV infected cell culture lysate and vero cells (as negative control) was spotted on NCM and air dried for 10 minutes, followed by blocking of NCM using the 5% Skim Milk Powder (SMP) in 0.05% Tween-20. Then the membrane was incubated with mAbs (1 mg/ml) at 1 minute and 5 minute. This step was followed by incubation with secondary antibody (1:1000 dilution of Anti-mouse conjugate, Sigma), diluted in blocking buffer. The membrane was washed three times in between each step with PBS+0.05% Tween-20. Finally, the Diaminobenzidine (DAB) was added as substrate and the distilled water was used to stop the reaction. The result was interpreted based on the intensity of color developed (as brown spots) in the membrane.
Indirect ELISA was used to study the interaction kinetics of both the mAbs i.e. the reactivity of mAbs against CDV antigen at different time intervals. The reference standard antigens, ultra purified CDV, PEG concentrated CDV and CDV infected Vero cell culture were used for coating the ELISA micro plates/modules at 37℃ for 1 hour (hr). Uninfected vero cell culture lysate was used as negative control. The micro plates were washed three times using PBS+0.05 % Tween-20, each mAb (1 mg/ml) was added to respective set and the plates were incubated for 1 minute (min), 5 min, 10 min, 30 min, and 1 hr. After washing the plates, the secondary antibody (1:1000 dilution of antimouse conjugate, Sigma) was added and incubated for 1 hour. The micro plates were washed and the O Phenylenediamine Dihydrochloride (OPD) was used as substrate. After the development of color, the reaction was stopped by adding 1 M H2SO4. The absorbance or optical intensity was taken in ELISA reader at 492 nm.
Assembly of lateral flow assay strip
LFA strips were assembled following the method of Sajid, et al.[13]. For the spotting on the test and control line, anti-CDV mAb and antimouse IgG (whole molecule) (Sigma M7023) were used, respectively. Spotting was done on NCM laminate of pore size (10 μm) using the easy printer. The laminate was kept at room temperature for 1-2 hrs to dry. The sample pad, conjugate pad and absorption pad were later attached to the NCM laminate with a 1-2 mm overlap. The LFA strip was cut manually (5 mm each) and stored at 4°C until use.
Preparation of gold nanoparticle-mAb bio conjugate
NaCl flocculation test was done primarily to check the stability of 30 nm Gold Nanoparticles (GNP) at different concentrations of mAb (10 μg/ml, 20 μg/ml, 30 μg/ml) and different pH of buffer (7.5, 8.5, 9.5).
Bioconjugate was prepared by the physical adsorption method, with some modifications as per the requirement of specific mAb [14]. The optimized concentration of mAb was mixed with 5 mM KH2PO4 (pH 7.5) to make the solution 100 μl. 900 μl of GNP was added to the solution and incubated at room temperature for 30 min. The mAb-GNP bio conjugate formed by physical adsorption during the incubation was blocked and stabilized by the addition of 100 μl of 10% Bovine serum albumin (BSA) (prepared in 50 mM KH2PO4, pH 9.0) and 50 μl of 1% Polyethylene glycol (PEG) (prepared in 50 mM KH2PO4, pH 9.0, pH 7.5), respectively. The suspension was again incubated at room temperature for 30 min and then centrifuged at 14,000 rpm for 20 min at 4℃. The supernatant was discarded and the pellet was dissolved in 1 ml of preservation solution (20 mM Tris HCl, 0.1% BSA, 0.01% sodium azide, 0.05% PEG-20,000, pH 8.2). The suspension was incubated at room temperature for 1 hr, centrifuged under the same conditions and the final pellet of bio conjugate was re suspended in 20-30 μl of remaining preservation solution. The bio conjugate was stored at 4°C until use.
Selection of combination of capture (test line) and detecting (bio conjugate) antibody
After screening the interaction kinetics in iELISA and binding affinity in NCM by dot blot assay, the final screening was done in LFA. All the four possible combinations using both the mAbs were screened in LFA using CDV antigen as reference standard along with respective negative control antigen. The reference standard antigens included in the study are CDV infected vero cell culture (106.5 TCID50/ml), 10 times PEG concentrated CDV antigen (~107.5 TCID50/ml) and the ultrapurified CDV.
Optimization of running buffer
Primarily, Phosphate Buffer Saline (PBS) at different pH ranges (7, 7.5, 8, and 8.5) was screened to obtain an efficient flow of sample and clear binding of bio conjugate. Further, Tween-20 and Triton X-100 at different concentrations (0.5%, 0.2%, 1%, 2%, 4% and 5%) were added to the PBS. To enhance antigen availability and increase sensitivity of the assay, some buffers to increase lyses of cell and virus were also screened (Table 1).
| S.no. | Buffer | Composition | Function |
|---|---|---|---|
| 1 | Sample running buffer | PBS+5% Triton-X, pH 8.0 | Provide optimum viscosity and cell lysis |
| 2 | RIPA cell lysis buffer | 10 mM Tris-HCl, pH 8.0 + 1 mM EDTA + 0.5 mM EGTA +1% Triton X-100 + 0.1% Sodium Deoxycholate + 0.1% SDS + 140 mM NaCl + 1 mM PMSF before use | Disrupt nuclear membranes and solubilize cytoplasmic proteins, resulting in a high protein yield. |
| 3 | Antigen extraction buffer 1 | PBS + 0.1%SDS + 0.05% Tween-20 | Virus lysis |
| 4 | Antigen extraction buffer 2 | 0.5 MTris + 0.1 MNaCl + 2% SDS + SMP + 0.1 m EDTA | Virus lysis |
Table 1: List of different running buffers screened for the enhancement of sensitivity of the LFA.
Assay procedure
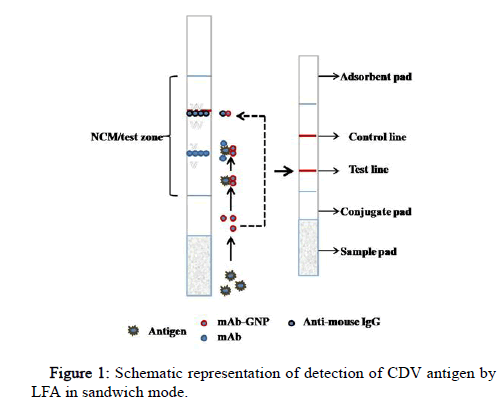
The sample and the running buffer were mixed in an Eppendorf tube, the GNP probe/bio conjugate was added to the conjugate pad and the LFA strip was dipped (dipstick method). This allows the analyte in the sample to conjugate with a primary/capture antibody labeled with gold nanoparticle (bio conjugate) on the conjugate pad. Analyze bio conjugate complex then flows by capillary action to NCM spotted with detecting antibody (test line) and secondary antibody (control line). The secondary antibody is specific to the species in which the primary antibody (mAb) is raised. The red color band at both the test and control line indicates a positive sample, and the red band only at the control line indicates a negative (Figure 1).
Analytical performance characteristics
The LFA’s analytical sensitivity was determined by using a 2 fold serially diluted suspension of PEG concentrated CDV antigen in a negative swab sample. The limit of detection was determined based on the signal intensity at the test line. The highest dilution which produced a visible red color band at the test line and control line was considered the cut-off point of the assay.
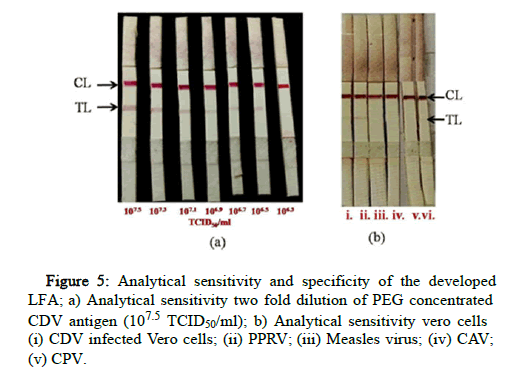
To test the specificity of the LFA, cell culture supernatant of Peste des Petits Virus (PPRV), Measles virus, Canine Adenovirus (CAV), and Canine Parvovirus (CPV) were screened.
Diagnostic performance characteristics
To test the analyte in its biological matrix, spiking of known negative clinical samples available in the laboratory repository was done. For spiking, 10% suspension of tissue homogenates (brain and lymph node), swab materials (nasal, ocular, and rectal swabs), blood and serum of dogs and different wildlife animal’s origin were used. These clinical samples were spiked with ultra-purified CDV antigen in a 1:1 ratio and the Vero cells were added (1:1) in unspiked clinical samples, as a negative control antigen. The samples were kept at 37℃ for 1 h allowing the sample matrix to equilibrate [15]. The spiked samples were used as true positive and the unspiked as true negative representatives.
The clinical samples included in the study were available in the laboratory repository and some of the samples were collected from the institutes veterinary polyclinics during the period of study. The samples were screened by the optimized LFA strips as well as by N gene based RT-PCR [16]. Diagnostic sensitivity and diagnostic specificity were determined using a two sided contingency table by comparing the positive and negative clinical samples tested with RTPCR and LFA.
Repeatability of the assay
The purified mAb was stored at -20℃ and used for spotting the laminates in 1 month intervals to study the stability of mAb. Also, the spotted strips were stored at 4℃ as well as room temperature to check the stability of mAb in the LFA strips.
Applicability of the LFA strip as sample substitute
Positive LFA strips were kept at room temperature as well as 4℃ for 7 days to determine the stability of absorbed CDV antigen for nucleic acid detection. Few modifications were added to the Trizol RNA extraction protocol and cDNA synthesis was as per the standardized protocol of the available kit (Quantitect Reverse Transcription kit cDNA synthesis (Qiagen, 205311). The N gene and H gene (in house developed primers) fragments were amplified to evaluate the applicability of LFA strips as sample substitute for molecular study in the laboratory.
Results
Up scaling and purification of monoclonal Antibodies (mAbs)
Hybridoma clones were collected at log phase by centrifugation at 2000 rpm for 5 minutes. The pellet was re suspended in serum free media, and inoculated in mice intra peritoneally after two days of IFA inoculation. The ascites fluid was collected between the 7th to 12th day of inoculation using an 18 gauge needle when sufficient abdominal distension was noticed. A total of 8-10 ml of ascites was collected for both the mAbs. Based on the iELISA, the reactivity of the mAbs in ascites fluid (1:32,768 titre) was found to be approximately 1000 times more than in hybridoma culture supernatant (1:32 titer). The ascites fluid was subjected to purification and the eluted antibodies were dialyzed to remove the salts of elution buffer, then re suspended and concentrated. The reactivity of the mAbs was confirmed and the titre was found to be nearly equal to that of ascites fluid.
Antigen-antibody interaction
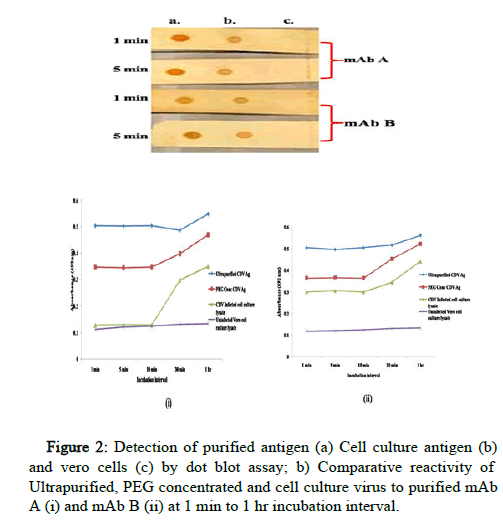
The reactivity of both the mAbs was checked in NCM to mimic the matrix as well as assay run time of LFA. Both the mAbs were found to possess high binding affinity against CDV antigen at both 1 min and 5 min incubation. The binding of both the mAbs with ultra-purified antigen was comparatively stronger than that of cell culture infected lysate (Figure 2).
The interaction kinetics of both the mAbs was also studied by iELISA against three different CDV antigen reference standards. Ultra purified and PEG concentrated CDV antigen were found to be reactive to both the antibodies even at 1 min incubation and the intensity increases with increased incubation time. The mAb B shows reactivity to cell culture infected lysate of CDV antigen at 1 min. mAb A was found to be comparatively low in binding against CDV infected Vero cell culture from 1 to 10 minutes and showed reactivity after 30 min incubation as depicted in (Figure 2). The reactivity in negative control was negligible and clearly differentiated from the positive controls.
Assembly of lateral flow assay strip
The test strip was assembled using its different components as shown in Figure 1. The spotting of the control and test line was done using the optimized concentrations of anti-mouse IgG and mAb, respectively. The distance of the test line was kept at 0.8 cm from the conjugate pad and 1 cm to the control line. The assembled LFA strips were stored at 4℃.
Preparation of gold nanoparticle-mAb bio conjugate
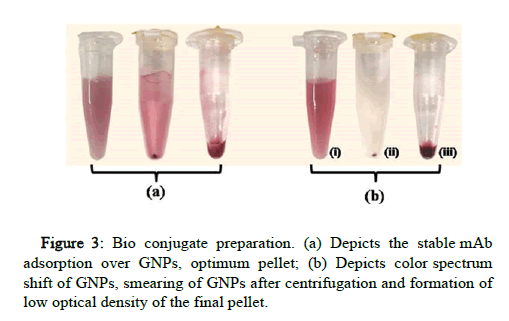
10 μg/ml of mAb B and 15 μg/ml of mAb A were found to be the most optimum concentration for stable bio conjugation of GNPs at pH 7.5. Both the mAbs were separately optimized with specific concentration and pH conditions. The bio conjugates were stable at 4℃ for a month without the settlement of GNP aggregates (Figure 3). On testing the strips, the bio conjugate was released from the conjugate pad with cleared NCM indicating the stable adsorption of mAb onto GNP.
With the lower concentration of mAb and higher pH of the buffers, the GNP color was shifted towards blue, indicating aggregation of GNPs which later results in the smearing of the pellet after centrifugation and low optical density of final bio conjugate. The unstable bio conjugate also results in higher non-specific binding at the test line.
Different volumes of bio conjugate were placed on the conjugate pad of the LFA strip and dipped in an eppendorf tube containing 100 μl running buffer. The 2 μl volume of bio conjugate showed intense binding at the control line without any smearing of GNP in test zone/ NCM. The control line forms by the binding of bio conjugate with anti-mouse IgG indicating the optimum function of bio conjugate.
Selection of combination of capture (test line) and detecting (bio conjugate) antibody
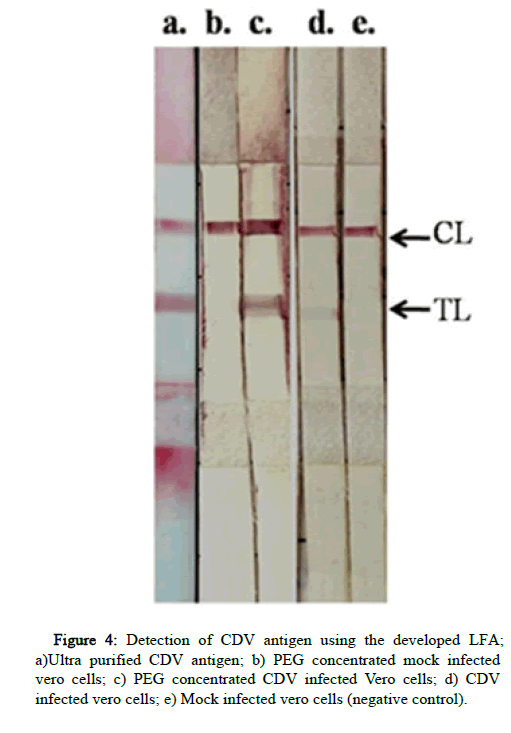
Different concentrations of both the mAbs such as 1 mg/ml, 2.5 mg/ml, and 5 mg/ml were tested for each antibody. At these concentrations no binding was observed, therefore the highest concentrations of antibodies obtained (13 mg/ml of mAb A and 6 mg/ml of mAb B) were used for the test line. All the four combinations of antibody pairs in sandwich format of LFA were tested. The mAb B at test line as well as bio conjugate showed high non-specific binding to uninfected vero cells. The mAb A was found to be suitable with optimal binding of CDV antigen and least nonspecific binding. All the three reference standard CDV antigens were detected (Figure 4). The higher intensity observed in PEG concentrated antigen than ultra purified antigen, was found to be due to the high viscosity of the PEG suspension rather than its antigenic mass. Therefore, the mAb a pair as test line as well as bio conjugate was finalized to develop LFA strips for antigen detection.
Optimization of running buffer
The running buffer mixed with the sample before running in the strip plays an important role in providing adequate viscosity to the sample, clearance in the NCM, etc. Some buffers were used as the medium to lyse the virus particles and enhance the availability of the intra vision antigen, but no improvement in sensitivity was observed (Table 1). Therefore, the 0.1 mM PBS+x% Triton-x 100, pH 8.0 was selected as sample running buffer.
Diagnostic efficacy of assay
Positive and negative control samples were mixed with running buffer at the ratio of 1:1 and 2 μl of bio conjugate was applied to the conjugate pad. The optimum assay run time was found to be around 3-5 min. The known positive CDV antigens showed two bands in the strip with only one band in negative control.
Analytical sensitivity and specificity
2-fold dilution of PEG concentrated CDV antigen was used to determine the detection limit of the assay. The dilutions were made in known negative nasal swab samples to ensure the suitability of the matrix. The limit of detection was found to be low, i.e. 106.5 TCID50/ml titre i.e only up to 3,162,277 virus particles were detected (Figure 5). No cross reactivity was observed with PPRV, Measles virus, Canine Adenovirus, Canine Parvovirus.
Diagnostic sensitivity and specificity
Developed LFA differentiated the spiked and unspiked samples, indicating its suitability in different matrices. Clinical samples including blood, serum, swabs and tissue homogenates were screened by the LFA as well as RT-PCR. The detection was more accurate in freshly collected samples from the clinic as compared to the stored ones, indicating the presence of high titre of virus in fresh samples. The diagnostic sensitivity and specificity was found to be 70.5% and 100%, respectively based on the result of LFA and RT-PCR of clinical samples from domestic dogs as well as different wildlife animal species. A substantial agreement between LFA and RT-PCR was observed as reflected by the kappa value of 0.756 with 95% confidence interval.
Repeatability of the LFA
The purified mAb stored at -20℃ was stable for 3 months and beyond it reduction in binding intensity was observed. The spotted strips were stable for more than 3 months at 4℃ and the repeatability of the results was observed. At room temperature (28-35℃), reduction in binding intensity of CDV antigen was observed.
Utilization of the LFA strip as sample substitute
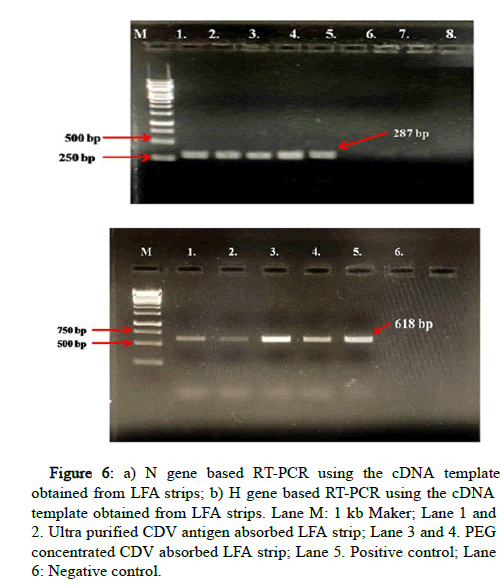
The positive LFA strips were kept at room temperature as well as at 4℃ for 1 week and then to study the stability of adsorbed CDV antigen in the LFA strips. The strips of both the sets then were subjected to RNA extraction. No amplification was observed in the strips stored at room temperature, indicating the instability of adsorbed CDV antigen at room temperature. The strips stored at 4℃ showed amplification but the intensity was lower than the LFA strip immediately processed. The positive cDNAs prepared from LFA strips of highly concentrated CDV antigens were used for amplification of N gene and H gene fragment (Figure 6). The study indicates that the LFA strips can be used as wet sample substitute for molecular studies, given there is high concentration of antigen in the samples.
Figure 6: a) N gene based RT-PCR using the cDNA template obtained from LFA strips; b) H gene based RT-PCR using the cDNA template obtained from LFA strips. Lane M: 1 kb Maker; Lane 1 and 2. Ultra purified CDV antigen absorbed LFA strip; Lane 3 and 4. PEG concentrated CDV absorbed LFA strip; Lane 5. Positive control; Lane 6: Negative control.
Discussion
CDV is an emerging pathogen and rising reports of infection in a number of wildlife animal species is a major concern. Twenty lineages have been reported till today indicating the continuous emergence and recombination of the virus. Dogs are the primary host of CD and play a key role in the transmission of the virus to these wildlife species. Therefore the prime step to control the disease is to arrest the virus circulation in dogs, which needs the use of accurate diagnostics and vaccines. Accurate diagnostics means targeting the circulating strains specifically, to avoid any false negatives. The present study aims the development of LFA for the detection of circulating CDV strains in the dog as well as wildlife population.
The intended purpose of the assay development for antigen detection is a confirmatory diagnosis of CDV clinical or suspect cases. The detection limit of the developed assay was found to be low (106.5 TCID50/ml) i.e minimum of 31 lakh vision particles in a sample. Therefore, few buffers were screened to enhance the sensitivity of the assay by increasing the availability of antigen. Detergents like Sodium deoxycholate, Triton-X 100, Tween-20, etc. are reported to be effective for the separation of membrane proteins for sensitive detection in an immune assay [17,18]. With Triton-X, the highest reduction in non-specific binding at test line and the optimum flow rate was found, whereas no improved effect in antigen binding was found. Cell culture lysates were used after three cycles of freeze thaw to ensure the lyses of cells to expose the virus particles. To release the intra vision N protein, viral lysis buffers or extraction buffers were also screened [19]. The non-specific binding at the test line was consistently found even with buffers due to additional reagents like NaCl, EDTA, etc. Also, no improvement in the test line intensity of positive samples was found, therefore the sample running buffer was kept at minimum salt composition. The high concentration of mAbs at the test line might be the reason for the physical adsorption of different components resulting into non-specific binding.
Rapid diagnostic tests against the Influenza virus developed using N protein presents the same limitation of lower detection limit as compared to the one based on haemagglutinin and fusion protein [20]. LFA based on mAbs against the surface proteins, H and F of PPRV has been found to be specific for clinical detection, whereas the sensitivity has been a constant issue in many studies [21-23]. A CDVLFA was also successfully developed using two monoclonal antibodies (mAbs) of IgG1 isotope against two different epitopes of the surface protein F resulting in sensitive as well as specific detection [24]. But there are few studies reporting the highly sensitive as well as specific antigen detection by LFA targeting intra vision proteins as well. In some studies, N protein has been targeted in LFA for the detection of viruses with similar vision structures like Influenza, PPRV, SARS-CoV-2, etc. SARS-CoV LFA targeting N protein is been reported to be highly specific as well as sensitive [25]. A study involving mAbs against both H and N proteins of PPRV has been conducted by our laboratory and both the mAbs were found to be suitable for antigen detection in LFA format with significant sensitivity (unpublished). Therefore it can be concluded that the binding affinity of the mAb in LFA format is equally important as its specificity to the target protein or epitope.
Additionally, the characterization of biological activity as well as physico-chemical properties of mAb is critical part that should be considered. Any kind of chemical or physical instabilities during the bio conjugate preparation or purification of mAb may lead to reduction in its binding affinity and the efficacy of the assay [26]. The study of physical properties like isoelectric point and affinity constant can be helpful in optimizing the buffers used in the study to provide maximum stability and biological potency to the mAbs. A negative charge causes a repulsion of the particles and prevents the optimum activity of antibody during bio conjugate preparation [27]. A stable bio conjugate formed by the optimum physical adsorption of mAb and GNP shows the less non-specific binding, which appreciates the detection of low quantity of analyte in the sample. Unstable and aggregated bio conjugate shows high non-specific binding than the freshly prepared stable bio conjugate [28].
Conclusion
Therefore we conclude that the assay can detect specifically the CDV antigen in freshly collected samples having high titre of the virus, validating the efficiency of assay to work in field conditions.
LFA provides very less interaction time for antigen and antibody interaction, therefore demands mAbs with high binding affinity. Therefore, to further enhance the sensitivity of LFA, the other mAb having higher affinity in LFA format or the mAbs against surface epitopes or proteins (H or F) can be screened.
Acknowledgement
This study is part of PhD thesis work of the first author. The authors are thankful to the Director and Joint Directors ICAR-IVRI for providing the necessary facility. The authors would like to acknowledge the funding agency Science and Engineering Research Board Department of Science and Technology (SERB-DST) Government of India (Grant no EEQ/2018/000823).
Conflict of Interest
The authors declared no potential conflicts of interest concerning research, authorship, and/or publication of this article.
References
- Loots AK, Mitchell E, Dalton DE, Kotzé A, Venter EH (2017) Advances in Canine Distemper Virus (CDV) pathogenesis research: a wildlife perspective. J Gen Virol 98: 311–321.
[Crossref] [Googlescholar] [Indexed]
- Lunardi M, Darold GM, Amude AM, Headley SA, Sonne L, et al. (2018) Canine distemper virus active infection in order Pilosa, family Myrmecophagidae, species Tamandua tetradactyla. Vet Microbiol 220: 7-11.
- Rendon-Marin S, Budaszewski RF, Canal CW, Ruiz-Saenz J (2019) Tropism and molecular pathogenesis of canine distemper virus. Virol J 16: 30.
- Beineke A, Puff C, Seehusen F, Baumgärtner W (2009) Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet Immunol Immunopathol 127:1-18.
- Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, et al. (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46:708-717.
- Murphy FA, Gibbs EPJ, Horzinek MC, Studdert MJ (1999) Veterinary Virology. 3rd (ed). Academic Press, San Diego, California, pp.629.
- Duque-Valencia J, Forero-Muñoz NR, Díaz FJ, Martins E, Barato P, et al. (2019) Phylogenetic evidence of the intercontinental circulation of a Canine distemper virus lineage in the Americas. Sci rep 9(1): 1-15.
- Bhatt M, Rajak KK, Chakravarti S, Yadav AK, Kumar A, et al. (2019) Phylogenetic analysis of haemagglutinin gene deciphering a new genetically distinct lineage of canine distemper virus circulating among domestic dogs in India. Transbound Emerg Dis 66:1252-1267.
- Rendon-Marin S, Martinez-Gutierrez M, Suarez JA, Ruiz-Saenz J (2020) Canine Distemper Virus (CDV) Transit Through the Americas: Need to Assess the Impact of CDV Infection on Species Conservation. Front Microbiol 11: 810.
- Wang R, Wang X, Zhai J, Zhang P, Irwin DM, et al. (2021) A new canine distemper virus lineage identified from red pandas in China. Transbound Emerg Dis 69:944-952.
- Hristov DR, Rodriguez-Quijada C, Gomez-Marquez J, Hamad-Schifferli K (2019) Designing paper-based immunoassays for biomedical applications. Sensors 19: 554.
- Hendriksen CFM, De Leeuw W (1998) Production of monoclonal antibodies by the ascites method in laboratory animals. Res Immunol 149:535-542.
- Sajid M, Kawde AN, Daud M (2015) Designs, formats and applications of lateral flow assay: A literature review. J Saudi Chem Soc 19:689-705.
- Kamel M, Salah F, Demerdash Z, Maher S, Atta S, et al. (2019) Development of new lateral-flow immunochromatographic strip using colloidal gold and mesoporous silica nanoparticles for rapid diagnosis of active schistosomiasis. Asian Pac J Trop Biomed 9: 315.
- Lugos MD, Damulak OD, Perikala V, Davou GI, Obeta UM, et al. (2019) Assay linearity and spike-recovery assessment in optimization protocol for the analysis of serum cytokines by sandwich ELISA platform. Am J Biomed Sci 3.
- Frisk AL, König M, Moritz A, Baumgärtner W (1999) Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J Clin Microbiol 37:3634-3643.
- Salmi A, Lund G (1984) Immunoassays for measles virus nucleocapsid antigen: effect of antigen-antibody complexes. J Gen Virol 65(10): 1655-1663.
- Geumann C, Gronborg M, Hellwig M, Martens H, Jahn R (2010) A sandwich enzyme-linked immunosorbent assay for the quantification of insoluble membrane and scaffold proteins. Ana Biochem 402:161-169.
- Grant BD, Anderson CE, Alonzo LF, Garing SH, Williford JR, et al. (2021) A SARS-CoV-2 coronavirus nucleocapsid protein antigen-detecting lateral flow assay. PloS one 16: 0258819.
[Crossref] [Googlescholar] [Indexed]
- Nguyen HP, Kwak C, Heo CK, Cho EW, Yang J, et al. (2018) Development and characterization of monoclonal antibodies against nucleoprotein for diagnosis of influenza A virus. J Microbiol Biotechnol 28: 809-815.
- Brüning-Richardson A, Akerblom L, Klingeborn B, Anderson J (2011) Improvement and development of rapid chromatographic strip-tests for the diagnosis of rinderpest and peste des petits ruminants viruses. J Virol Met 174:42-46.
- Baron J, Fishbourne E, Couacy‐Hyman E, Abubakar M, Jones BA, et al. (2014) Development and testing of a field diagnostic assay for peste des petits ruminants virus. Transbound Emerg Dis 61:390-396.
[Crossref] [Googlescholar][Indexed]
- Kinimi E, Odongo S, Muyldermans S, Kock R, Misinzo G (2020) Paradigm shift in the diagnosis of peste des petits ruminants: scoping review. Acta Vet Scand 62:1-14.
- An DJ, Kim TY, Song DS, Kang BK, Park BK (2008) An immunochromatography assay for rapid antemortem diagnosis of dogs suspected to have canine distemper. J Virol Methods 147: 244–249.
[Crossref] [Googlescholar][Indexed]
- Diao B, Wen K, Zhang J, Chen J, Han C, et al. (2021) Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect 27: 289-291.
- Le Basle Y, Chennell P, Tokhadze N, Astier A, Sautou V (2020) Physicochemical stability of monoclonal antibodies: a review. J Pharma Sci 109:169-190.
- Correro MR, Sykora S, Corvini PFX, Shahgaldian (2017) Enzyme Armoring by an Organosilica Layer: Synthesis and Characterization of Hybrid Organic/Inorganic Nanobiocatalysts. Methods Enzymol 590: 77-91.
- Liu Y, Zhan L, Qin Z, Sackrison J, Bischof JC (2021) Ultrasensitive and highly specific lateral flow assays for point-of-care diagnosis. ACS nano, 15:3593-3611.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi