Research Article, J Mol Biol Methods Vol: 7 Issue: 1
Intravenous Delivery of a Dicer Substrate siRNA to the Central Nervous System via Rabies Virus Glycoprotein
Ashley Henderson1, Patricia Mott2, Tamara Morris1, Sarah Melton1, Peter Winsauer1,2, and Ashok Aiyar2*
1Department of Pharmacology and Experimental Therapeutics, Louisiana State University Health Sciences Center, New Orleans, LA, United States of America
2Department of Biochemistry and Molecular Biology, Louisiana State University Health Sciences Center, New Orleans, LA 70112, United States of America
*Corresponding Author: Ashok Aiyar,
Department of Biochemistry and Molecular Biology,
Louisiana State University Health Sciences Center, New Orleans, LA 70112, United States of
America
E-mail: aaiyar@lsuhsc.edu
Received date: 21 March, 2024, Manuscript No. JMBM-24-130399;
Editor assigned date: 25 March, 2024, PreQC No. JMBM-24-130399 (PQ);
Reviewed date: 08 April, 2024, QC No. JMBM-24-130399;
Revised date: 15 April, 2024, Manuscript No. JMBM-24-130399 (R);
Published date: 22 April, 2024, DOI: 10.4172/JMBM.1000147
Citation: Henderson A, Mott P, Morris T, Melton S, Winsauer P, et al. (2024) Intravenous Delivery of a Dicer Substrate siRNA to the Central Nervous System via Rabies Virus Glycoprotein. J Mol Biol Methods 7:1.
Abstract
Introduction: A Rabies Virus Glycoprotein (RVG) peptide was found to be an effective carrier for siRNA molecules across the selectively impermeable blood brain barrier and into neurons. This technique facilitates intravenous small interfering RNA administration, but its application remains limited to mice. Expansion into other animal species is required to broaden its research use and enhance its clinical relevance.
Methods: The RVG peptide was fused with an alternating arginine-glycine tail and separated by a glycine spacer. This peptide component was then combined with a Dicer substrate small interfering RNA (DsiRNA) designed to knock down mRNA specific to a protein, Shisa7, that associates with GABAA receptors. Two cohorts of rats implanted with femoral catheters were dosed once per day for two consecutive days and sacrificed at varying time points after administration of RVG-9r/DsiRNA. Brain tissue was analyzed by RT-PCR and Western blot analysis to verify gene silencing.
Results: The first cohort of rats was analyzed via Western blot analysis. The pons/medulla and cerebellum showed robust knockdown of Shisa7, with a mean reduction of 51% and 68%, respectively. Shisa7 in the frontal cortex was reduced by 43% and 6% at the 24-hour and 48-hour time point, respectively. The second cohort of rats produced similar results for the pons/medulla and cerebellum, while the frontal cortex showed a reduction of 17% and 49% at 24 and 48 hours. The hippocampus only showed gene silencing at 24 hours post-infusion. The RT-PCR data supported the Western blot data, in that Shisa7 mRNA levels decreased in multiple brain areas.
Conclusions: This study effectively knocked down Shisa7, an auxiliary protein of the GABAA receptor complex, in multiple brain areas after intravenous delivery of RVG-9r/DsiRNA in rats, making this non-invasive technique a viable method for studying Central Nervous System (CNS) proteins.
Keywords: Rabies virus glycoprotein; Dicer substrate small interfering
RNA; Quantitative PCR; Western blot; ?-aminobutyric acid type A receptor.
Abbreviations
Central Nervous System (CNS), γ-Aminobutyric Acid type A Receptor (GABAAR), Dicer substrate small interfering Ribonucleoic Acid (DsiRNA), Rabies Virus Glycoprotein (RVG), Intracerebroventricular (ICV)
Introduction
Studying protein function poses significant challenges because of their complex interactions with other proteins and receptors. Two of the most common methods for studying proteins are by germline knockout or protein knockdown, which reduce or suppress a specific protein’s expression or activity within a cell or organism. These procedures are invaluable because the absence or reduction of a protein can provide important information about the processes or functions in the organism that are reliant on that protein [1]. However, gaining access to CNS proteins can be difficult due to the relatively impermeable blood brain barrier. Intraparenchymal and intracerebroventricular injections place solutions directly into the brain or surrounding cerebrospinal fluid, though these injections can cause varying degrees of brain trauma [2]. Intraparenchymal injections affect a small, targeted area of the brain, while intracerebroventricular (ICV) injections are used for more comprehensive knockdowns. One downside to ICV injections is low penetrance into brain tissue, so most of the affected tissue is located near the ventricles. An alternative method that has shown more extensive penetrance and widespread knockdown is tail-vein injection of adeno-associated viral vectors, which is often given with a mannitol pretreatment [3,4]. Another common approach to achieving protein knockdown is the use of small interfering RNA (siRNA), a member of a class of RNA molecules used for targeting and degrading messenger RNA (mRNA) [5]. The molecules are double stranded, usually 21 nucleotides in length, and use their complementary nucleotides to target specific mRNA. Once bound, they are incorporated into a protein complex called the RNA-Induced Silencing Complex (RISC), which facilitates mRNA degradation and prevents the translation of mRNA to protein. The effects of siRNA are usually rapid and transient depending upon any chemical modifications, with most siRNAs being degraded over the course of a few days [6].
Neurons can be particularly difficult to transfect with siRNA due to their complex and highly interconnected nature and the endothelial tight junctions of the blood brain barrier that prevent many molecules from entering the CNS [7]. Recently, Rabies Virus Glycoprotein (RVG) has gained significant attention in the field of neuroscience because it promotes the passage of the rabies virus across the blood brain barrier, into neurons, and from cell to cell [8]. RVG binds to nicotinic acetylcholine receptors in the brain, in addition to a number of other receptors from varying receptor families, including neural cell adhesion molecule, p75 neurotrophin receptor, metabotropic glutamate receptor subtype 2, and integrin β1, which mediate its entry into the cell [9-12]. The capacity of RVG to bind such a wide range of receptors is thought to contribute directly to the virus’ interspecies applicability and to making RVG an interesting protein for use in molecular biology [13]. Thus, multiple methods employing RVG have been used to allow siRNA to enter neurons, including RVG tagged exosomes and liposomes, RVG fused to enzymes, and RVG fused to an arginine tail that complexes with siRNA [14-17].
Of particular interest to us, was the method of fusing a 29-residue peptide derived from RVG (RVG-29) to a poly arginine peptide (9r) and complexing the positively charged tail with negatively charged siRNA in mice. This complex, when injected intravenously, showed efficient transfection with neuronal cell specificity [18]. RVG has been shown to bind the alpha-1 and alpha-7 subunits of the acetylcholine receptor and has also shown similarity to the snake alpha toxins [8,19]. By using the snake alpha toxin, α-Bungarotoxin (BTX), which binds to the alpha-7, -8, and -9 subunits of the acetylcholine receptor, Kumar et al. showed that their shortened RVG peptide competitively inhibited BTX’s binding to acetylcholine receptors [18,20]. The acetylcholine receptor appears to be responsible for RVG peptide uptake. They transfected mice with this complex and showed no changes in mRNA expression in the spleen or liver, but a 30% knockdown in gene expression in the brain. The peak knockdown was observed 2 days after their treatment regimen, with protein levels slowly increasing from that 48-hr time point. A different research team then lengthened the therapeutic timeline of the RVG-derived protein complex by developing a smaller RVG peptide (C2-9r) with more protection from serum nucleases [21]. In addition to shortening the peptide, they added a spacer of 4 glycine residues. These conformationally unrestrained amino acids provided more flexibility and promoted more efficient entry into the cell. The C2-9r/siRNA also showed neuronal specificity, with the largest knockdown occurring in the cerebellum and the least, but still significant, knockdown in the cortex and hippocampus.
In the present study, we combined the RVG-derived peptide from Kumar et al. and the 4-glycine spacer from Javed et al., with an arginine tail that was alternated with glycine residues [18,21]. This combination allowed the tail to bind RNA and DNA, similar to the protein, EBNA1, and its nucleic acid-binding motif [22]. Not only has the EBNA1 protein DNA-binding motif been shown to bind RNA, this same pattern of repeating glycine and arginine has been seen in a number of RNA-binding proteins [23,24]. Furthermore, we chose to use dicer substrate siRNA, which consists of 25-30 nucleotide double stranded RNAs that bind with Dicer endonuclease [25]. This modification makes DsiRNA 2-logs more effective at gene silencing than standard 21-mer siRNA [26]. DsiRNA was also shown to be less immunogenic than standard siRNA, which gives DsiRNA a better clinical applicability [25]. Early reports of in vivo delivery of DsiRNA have been promising and shown efficacious knockdown of various genes in peritoneal macrophages and the liver [27,28]. In contrast, knockdown for CNS-related purposes has had more limited success and requires direct injection or some type of carrier. Expanding this RVG-peptide guided siRNA to be less immunogenic DsiRNA and to a rat model would expand its use and promote investigation into its clinical usefulness. Analysis using NCBI’s BLAST on the percent identity between mouse and rat receptors known to be involved in rabies virus cellular uptake revealed >90% similarity between the two species (Table 1). This suggests the RVG-mediated delivery of siRNA may be translatable to a rat model. While previous work with RVGguided siRNA has only been accomplished in mice, we show through our work that the same method can be adapted to a rat model.
| Mouse | Rat | Percent identity | Mouse | Rat | Percent identity |
|---|---|---|---|---|---|
| Nicotinic Acetylcholine receptor subunit alpha 1 P04756.1 | Nicotinic Acetylcholine receptor subunit alpha precursor NP_077811.1 | 98.47% | Nicotinic Acetylcholine receptor subunit alpha 9 NP_001074573.1 | Nicotinic Acetylcholine receptor subunit alpha 9 AAA56720.1 | 97.49% |
| Nicotinic Acetylcholine receptor subunit alpha 1 P04756.1 | Nicotinic Acetylcholine receptor alpha polypeptide 1, isoform CRA_a EDL79151.1 | 97.84% | Nerve Growth Factor Receptor (TNFR superfamily, member 16) Q9Z0W1.2 | Tumor necrosis factor receptor superfamily member 16 P017174.1 | 95.78% |
| Nicotinic Acetylcholine receptor subunit alpha 7 AAF35885.1 | Nicotinic Acetylcholine receptor subunit alpha 7 precursor NP_036964.3 | 99.40% | Neural Cell adhesion molecule 1 P13595.3 | Neural cell adhesion molecule 1 isoform X8 XP_017450950.1 | 97.86% |
| Nicotinic Acetylcholine receptor subunit alpha 7 AAF35885.1 | Nicotinic Acetylcholine receptor subunit alpha 7 subunit splice variant 7-2 AAV31080.1 | 93.97% | Glutamate receptor, metabotropic 2 Q14B12.2 | Metabotropic glutamate receptor 2 precursor NP_001099181.1 | 98.51% |
| Nicotinic Acetylcholine receptor subunit alpha 9 NP_001074573.1 | Nicotinic Acetylcholine receptor subunit alpha 9 precursor NP_075219.2 | 97.70% | Integrin beta 1 (fibronectin receptor beta) P09055.1 | Integrin beta-1 P49134.1 | 98.12% |
Table 1: Mouse and rat proteins were compared using the NIH’s Basic Local Alignment Search Tool. Proteins were selected based on previous research showing interaction with rabies virus glycoprotein, suggesting possible involvement in RVG-9r-mediated cellular uptake of siRNA. High similarity between rat and mouse receptor proteins suggests that the RVG-9r/siRNA technique may be expanded from mice to rats.
Materials and Methods
Subjects and surgery
Long-Evans hooded rats, six male and one female, from Charles River (Wilmington, MA) and one Sprague Dawley male rat from Envigo (Indianapolis, IN) served as subjects in this experiment. The subjects’ diet consisted of 45-mg grain-based food pellets (Bio-Serv, Prospect, Connecticut) and Teklad 18% protein rodent diet chow (Indianapolis, IN). Starting at 6:00 am., the colony room was on a 14-hour/10-hour light/dark cycle and maintained at a temperature of 21°C ± 2°C with 50% ± 10% humidity. The subjects were individually housed in polypropylene cages with woodchip bedding and nesting material. All animals were handled in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Louisiana State University Health Sciences Center in New Orleans and the Guide for the Care and Use of Laboratory Animals. These subjects had a history of acute drug administration and of responding under operant schedules of reinforcement prior to this study.
To achieve a knockdown of Shisa7 using the RVG complex, the rats were implanted with a femoral catheter attached to a semipermanent exteriorized port located on subjects’ back that allowed for repeated acute i.v. infusions. Catheters remained patent for up to a year, with patency maintained by regular 1 ml/kg i.v. infusions containing heparin (50 U/ml), enrofloxacin (5 mg/ml), and 0.9% saline. Occasionally, catheter patency was assessed by i.v. administration of 0.1-0.2 ml of 10 mg/ml methohexital sodium (PAR Pharmaceuticals, Woodcliff Lake, New Jersey).
RVG-9r/DsiRNA
The RVG-9r with a 4-glycine spacer was obtained from Peptide 2.0 (Chantilly, VA) in a quantity of 30 mg. The full sequence was as follows: YTIWMPENPRPGTPCDIFTNSRGKRASNGG GGrGrGrGrGrGrGrGrGr, where r is the D form of arginine. It was stored in a 5% glucose solution and kept at -20°C. A pre-designed DsiRNA targeting rat Shisa7 (catalog # rn.Ri.Shisa7.13.1) was obtained from Integrated DNA Technologies (Coralville, IA).
RVG-9r/DsiRNA administration
Prior to infusion, a 1:1 molar ratio of RVG:9R peptide: DsiRNA at 3 nanomoles each was suspended in 200 microliters of 5% glucose solution. A 1:1 molar ratio was used, rather than the 10:1 molar ratio reported by Kumar et al. due to issues with precipitation upon mixing the 10:1 molar ratio [18]. We suspect precipitation of the 10:1 molar ratio was due to the excess of positively charged RVG peptide, which, when in the presence of a few molecules of negatively charged DsiRNA, bound and clumped together similar to what occurs in neutrophil extracellular traps [29]. Similar clumping was observed previously in studies evaluating the nucleic acid-binding domains of EBNA1 [30].
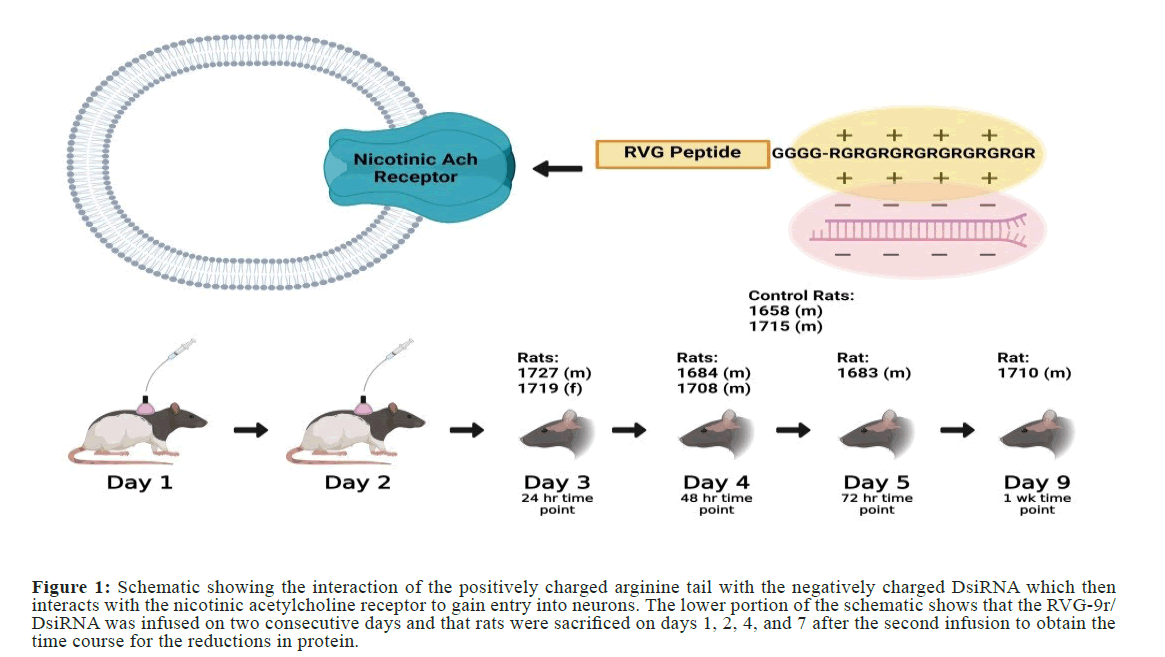
The entire 200 microliter solution was administered i.v. to the rats followed by a 200 microliter flush with a 5% glucose solution to clear any RVG-9R/DsiRNA from the in-dwelling portion of the catheter. The rats were dosed once per day for two consecutive days with the peptide/DsiRNA complex (3 nanomoles each) as an adaptation of the procedure used by Kumar et al. [18]. Rats were sacrificed at varying time points after the second dose of RVG-9r/DsiRNA (Figure 1). At these time points, the brains were extracted and dissected into separate regions using a previously established procedure [31]. The brain regions dissected were the frontal cortex, cerebellum, pons/ medulla, striatum, midbrain, hippocampus, and cortex. To ensure consistency across brain dissections, the same researcher completed every dissection, used specific gross features as a guide, and compared end weights and visual appearances of all brain areas between rats.
Figure 1: Schematic showing the interaction of the positively charged arginine tail with the negatively charged DsiRNA which then interacts with the nicotinic acetylcholine receptor to gain entry into neurons. The lower portion of the schematic shows that the RVG-9r/ DsiRNA was infused on two consecutive days and that rats were sacrificed on days 1, 2, 4, and 7 after the second infusion to obtain the time course for the reductions in protein.
Reverse-transcriptase-qPCR for Shisa7 gene expression
RT-PCR was performed on RNA isolated from a control rat and a rat that was given two consecutive days of RVG-9r/DsiRNA and sacrificed 48 hours after treatment. Following decapitation using a guillotine, the brains were extracted, dissected into the previously indicated brain areas, and flash frozen in liquid nitrogen. The samples were then stored in a -80°C freezer until further use. The subset of brain areas analyzed for qPCR were cerebellum, pons/ medulla, striatum, midbrain/thalamus, and hippocampus. RNA was extracted from 200 mg of rat brain tissue using TRIzol reagent per manufacturer’s protocol (Invitrogen). Extracted RNA was used to prepare cDNA using the iScript gDNA Clear cDNA Synthesis kit (Bio-Rad; catalog #1725035). qPCR assays were performed using the SsoAdvanced Universal SYBR Green Supermix kit (Bio- Rad; catalog #1725271). Reactions were run using Bio-Rad CFX 96. The following primers were used for amplification: Shisa7 Forward 5’-CCACCTGTTCCTGGGACCTCTC-3’ and Shisa7 Reverse 5’-TTCTCACGACATGGCAGAGA-3’. The mRNA levels were quantified relative to rat 18S mRNA levels using primers 18S Forward 5’-AAACGGCTACCACATCCAAG-3’ and 18S Reverse 5’TTGCCCTCCAATGGATCCT-3’ [32]. Both assays were performed at an annealing temperature of 53°C and melt curve analysis was used to confirm amplicon specificity.
Western blots
The first group of Western blots were performed with flash frozen tissue from dissected brain regions of Long-Evans rats treated with either control or RVG-9R/DsiRNA. The subset of brain areas analyzed for this analysis were cerebellum, pons/medulla, frontal cortex, and hippocampus. Of the treated rats, brains were obtained at 24 hours, 48 hours, and 72 hours. The protein extraction buffer contained 20 mM Tris pH 8.0, 137 mM NaCl, 0.5 mM sodium orthovanadate, 2 mM okadaic acid, 10% glycerol, 1% Nonidet P-40, 2% proteinase inhibitor based on a previously published procedure [33]. The antibodies used included rabbit polyclonal antibody Shisa7 (Biorbyt, Durham, North Carolina) (orb587814) as a primary antibody (1:200 dilution), antirabbit IgG, Horseradish Peroxidase-linked whole antibody (CiteAb, United Kingdom) (NA934) as a secondary antibody (1:1500 dilution), and HRP-conjugated actin mouse monoclonal antibody (Proteintech, Rosemont, Illinois) (HRP-60008) for detection of actin (1:10000 dilution). Dilutions and procedures for the Western blots were based on previously published procedures [34]. The Shisa7 protein band intensities were normalized to that of actin and the subsequent percent changes in protein levels of Shisa7 were graphed.
The second group of Western blots were completed with a different cohort of Long-Evans rats with the goal of replicating the data from the first group while also adding a one-week post-knockdown time point to assess protein recovery time. In these blots, the samples were also treated with TRIzol due to the need to simultaneously extract RNA from the samples. The protein extraction protocol was based on an optimized protein extraction buffer used for TRIzol treated samples [35]. All quantification was performed using Image J [36].
Results
RT-PCR
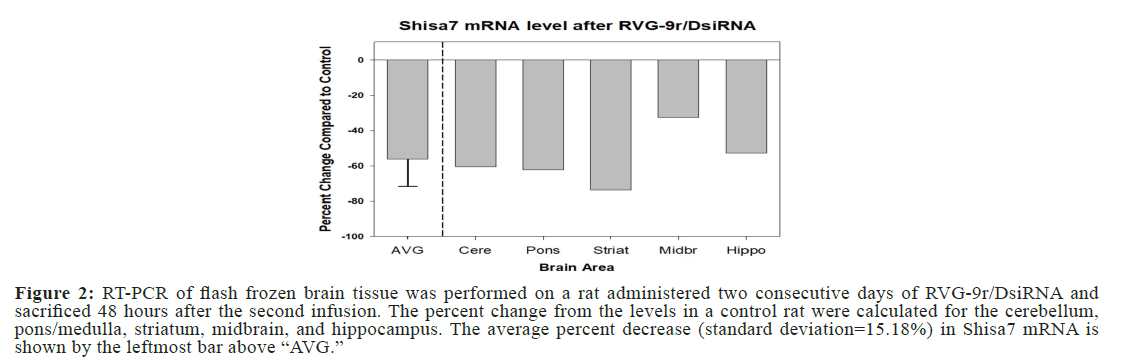
Comparison of Shisa7 mRNA levels between a control animal and an animal sacrificed at 48 hours after the second dose of the siRNA indicated Shisa7 mRNA levels decreased in multiple brain areas. More specifically, the reduction of Shisa7 mRNA levels was >50% in cerebellum, pons, striatum, and hippocampus, and 33% in midbrain. The average decrease of Shisa7 mRNA across all brain areas analyzed was 56% with a standard deviation of 0.15% (Figure 2).
Figure 2: RT-PCR of flash frozen brain tissue was performed on a rat administered two consecutive days of RVG-9r/DsiRNA and sacrificed 48 hours after the second infusion. The percent change from the levels in a control rat were calculated for the cerebellum, pons/medulla, striatum, midbrain, and hippocampus. The average percent decrease (standard deviation=15.18%) in Shisa7 mRNA is shown by the leftmost bar above “AVG.”
Western blots
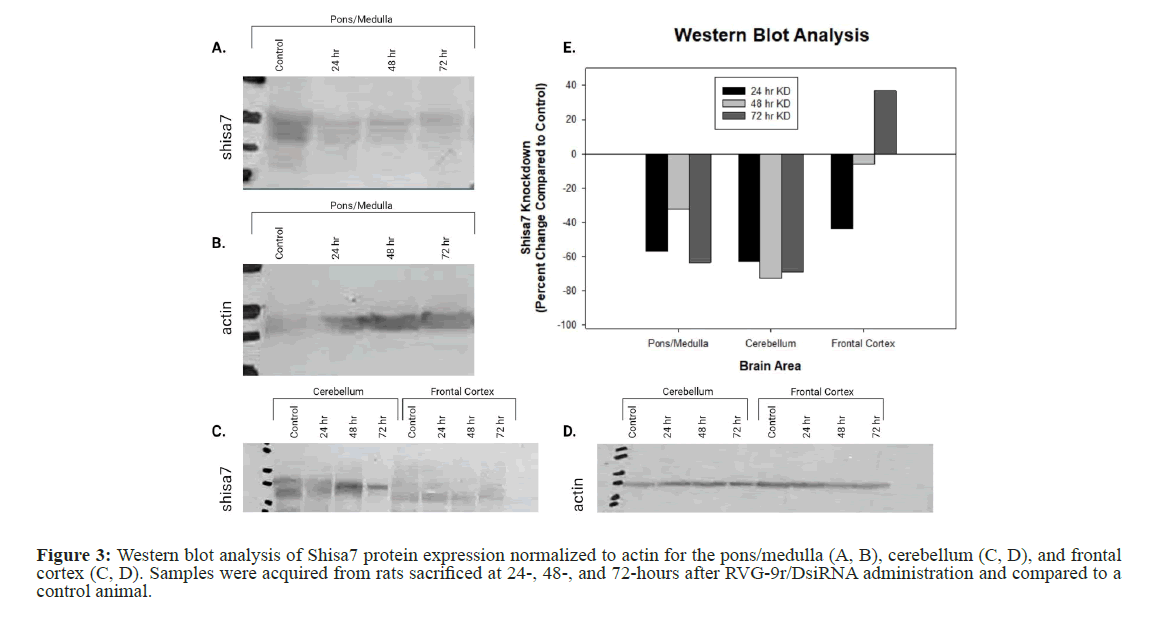
As shown in panels A-E of Figure 3, levels of Shisa7 were successfully reduced in the pons/medulla and cerebellum at all three time points, with the peak reductions varying between brain areas and across the time points for specific brain areas (pons/medulla 24 hr=-57.05%, 48 hr=-32.54%, 72 hr=-63.46%: Cerebellum 24 hr=- 62.71%, 48 hr=-72.81%, 72 hr=-69.18%: Frontal cortex 24 hr=-43.61, 48 hr=-5.96%, 72 hr=+36.91%). For example, a 32.54% reduction was obtained for the pons/medulla at the 48-hr time point, whereas a 62.71% reduction was obtained at the 72-hr time point. The frontal cortex samples showed a 43.61% reduction of Shisa7 at 24 hours after RVG-9R/DsiRNA administration, but protein appeared around control levels at the 48 and 72-hour time points (Figures 3C-3E).
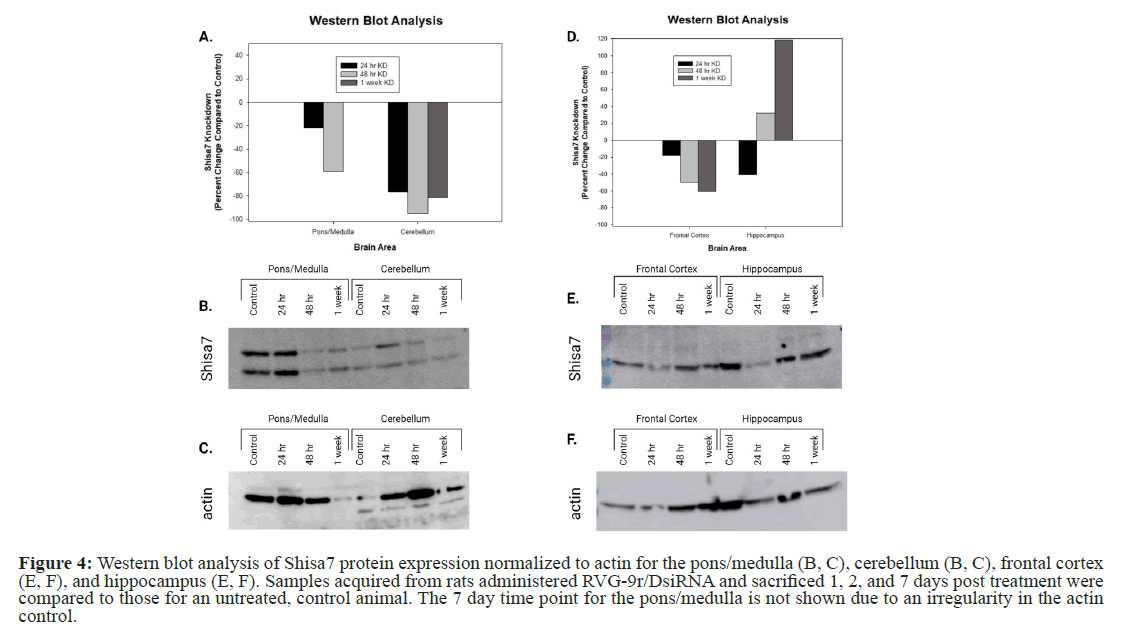
In the second cohort of rats that were administered RVG-9r/ DsiRNA and sacrificed at varying time points, similar results were seen compared to the initial cohort (pons/medulla 24 hr=-22.09%, 48 hr=-58.97%, 1 wk=+347% (excluded): Cerebellum 24 hr=-76.98%, 48 hr=-95.38%, 1 wk=-81.50%: frontal cortex 24 hr=-17.96%, 48 hr=-49.90%, 1 wk=-60.77%: Hippocampus 24 hr=-40.53%, 48 hr=+32.12%, 1 wk=+118.638%). The reduction of Shisa7 in the pons/medulla peaked at 48 hours, but the one-week time point was excluded from the analysis due to an irregular actin/Shisa7 ratio (Figure 4). There was also a marked reduction of Shisa7 in the cerebellum that persisted through the one-week time point (Figures 4A-4C), with a peak reduction of 95% after 48 hrs. Levels of Shisa7 in the frontal cortex were moderately reduced (50% at 48-hour time point) and this reduction persisted for one week. Unlike the other brain areas examined, Shisa7 in the hippocampus was only reduced at the 24-hr time point and protein levels were markedly upregulated at the 48-hr and one-week time points (Figures 4D-4F).
Figure 4: Western blot analysis of Shisa7 protein expression normalized to actin for the pons/medulla (B, C), cerebellum (B, C), frontal cortex (E, F), and hippocampus (E, F). Samples acquired from rats administered RVG-9r/DsiRNA and sacrificed 1, 2, and 7 days post treatment were compared to those for an untreated, control animal. The 7 day time point for the pons/medulla is not shown due to an irregularity in the actin control.
Discussion
The present study demonstrated the effectiveness of an RVGderived peptide fused to a 9-arginine tail with a 4-glycine spacer (RVG- 9r) for delivering dicer substrate small interfering RNA (DsiRNA) into the CNS of rats. Moreover, DsiRNA was successfully delivered into specific brain regions and reduced expression of Shisa7. The use of RVG-9r as a delivery vector has been previously demonstrated only in cell lines and mouse models, but our study extends this method to rats and offers advantages in terms of translational relevance [18]. Rats have been used extensively in behavioral and neuroscience research, making them a valuable species for investigating the effects of targeted protein reductions in a broader context. Not only does our study widen the scope of this application to an additional species, but it also employs the use of DsiRNA, which are shown to be 100 fold more efficient at reducing protein levels while also being less immunogenic than standard siRNA [25,26].
The choice of a 1:1 molar ratio of RVG-9r peptide to DsiRNA was selected after the 10:1 molar ratio reported in Kumar et al. created a precipitate prior to administering the solution i.v. [18]. We hypothesize that the excess of positively charged RVG peptide may have led to the formation of aggregates with the negatively charged DsiRNA, similar to the process seen in neutrophil extracellular traps [29,30]. There is also the possibility that the glycine spacer of the DsiRNA, which is longer than standard siRNA, was responsible for the precipitation at a 10:1 ratio. Nevertheless, the adjustment in ratio proved effective in preventing precipitation and allowed for successful delivery of DsiRNA into the rat brain.
The Western blot results from the first cohort of subjects confirmed a region-specific reduction of Shisa7 protein in the pons/ medulla and cerebellum, with a consistent reduction at 24, 48, and 72 hrs. The peak reduction of Shisa7 in these two brain areas varied, while the frontal cortex only showed a successful reduction after 24 hours. This temporal variation in kinetics suggests region-specific differences in RVG-9r-mediated DsiRNA delivery and cellular uptake, which may be due to differences in receptor expression, cellular uptake mechanisms in those brain regions, or differences in blood supply, considering the intravenous route of administration. To further elucidate the timespan over which the reduction in Shisa7 protein occurred and to validate the data from the first cohort of rats, we conducted a second set of Western blots using a second cohort of rats. In this cohort, the one-week time point in the pons/medulla had to be discarded due to irregularities in the actin/Shisa7 ratio, but the 24 and 48-hour time points both showed reductions in protein similar to that observed with the first cohort. The cerebellum also showed similar reductions of Shisa7 compared to the first group of rats, and this reduction persisted for seven days. The frontal cortex showed a modest reduction that also persisted for one week, which differed from the first group of rats where lower levels of Shisa7 only lasted 24 hours. Lastly, the hippocampus showed a brief reduction followed by Shisa7 protein upregulation, similar to the frontal cortex in the first cohort of rats. These data would seem to indicate that the reductions in Shisa7 were not equivalent across brain regions and that protein recovery also varied across the brain regions examined. This should not be surprising, however, as the robust reduction in the cerebellum and lower levels of protein in the cortex and hippocampus is similar to reductions reported previously when using a truncated RVG protein to carry siRNA, C2-9r/siRNA, into the brain [21].
Data obtained from RT-PCR analysis supports the observations seen at the protein level. The cerebellum, pons, striatum, midbrain, and hippocampus of a rat sacrificed 48 hours after administration of two doses of RVG-9r/DsiRNA showed reductions in Shisa7 mRNA compared to a control animal. This consistent pattern of Shisa7 protein reductions in addition to the decreased Shisa7 mRNA after i.v. RVG-9r/DsiRNA administration further supports the effectiveness of this delivery model and shows that it is translatable to a rat model.
Conclusion
The goal of the present study was to demonstrate reductions in Shisa7 as a proof-of-concept for the i.v. delivery of an RVG-9r/DsiRNA complex into the CNS of rats. Further, our findings demonstrated brain-area dependent reductions of Shisa7 and the differential recovery of Shisa7 in those areas over time. This knowledge provides researchers with the opportunity to directly assess the functional role of Shisa7 in those brain areas and its capacity to mediate the pharmacological response of those areas to both licit and illicit drug administration. Using this rat model to modulate protein levels could also be valuable for predicting the effects of drugs that are positive GABA (A) modulators (e.g., benzodiazepines), because Shisa7 has been shown to co-localize with GABA (A) receptors in the CNS.
References
- Alberts B (2002). Molecular biology of the cell 4th edition.
- Zhou K, Han J, Wang Y, Zhang Y, Zhu C (2022). Routes of administration for adeno-associated viruses carrying gene therapies for brain diseases. Front Mol Neurosci 15:988914.
- Fu H, Muenzer J, Samulski RJ, Breese G, Sifford J, et al. (2003). Self-complementary adeno-associated virus serotype 2 vector: Global distribution and broad dispersion of AAV-mediated transgene expression in mouse brain. Mol Ther 8(6):911-917.
- Hudry E, Andres-Mateos E, Lerner EP, Volak A, Cohen O, et al. (2018). Efficient gene transfer to the central nervous system by single-stranded Anc80L65. Mol Ther Methods Clin Dev 10:197-209.
- Alshaer W, Zureigat H, Al Karaki A, Al-Kadash A, Gharaibeh L, et al. (2021). siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur J Pharmacol 905:174178.
- Layzer JM, McCaffrey AP, Tanner AK, Huang ZA, Kay MA, et al. (2004). In vivo activity of nuclease-resistant siRNAs. RNA 10(5):766-771.
- Schlachetzki F, Zhang Y, Boado RJ, Pardridge WM (2004). Gene therapy of the brain: The trans-vascular approach. Neurology 62(8):1275-1281.
- Lian M, Hueffer K, Weltzin MM (2022). Interactions between the rabies virus and nicotinic acetylcholine receptors: A potential role in rabies virus induced behavior modifications. Heliyon 8(9).
- Thoulouze MI, Lafage M, Schachner M, Hartmann U, Cremer H, et al. (1998). The neural cell adhesion molecule is a receptor for rabies virus. J Virol 72(9):7181-7190.
- Tuffereau C, Bénéjean J, Blondel D, Kieffer B, Flamand A (1998). Low‐affinity nerve‐growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 17(24):7250-7259.
- Wang JinLiang WJ, Wang ZiLong WZ, Liu RenQiang LR, Shuai Lei SL, Wang XinXin WX, et al. (2018). Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog 14(7):e1007189.
- Shuai L, Wang J, Zhao D, Wen Z, Ge J, et al. (2020). Integrin β1 promotes peripheral entry by rabies virus. J Virol 94(2):10-128.
- Khalifa ME, Unterholzner L, Munir M (2021). Structural and evolutionary insights into the binding of host receptors by the rabies virus glycoprotein. Front Cell Infect Microbiol 11:736114.
- dos Santos Rodrigues B, Arora S, Kanekiyo T, Singh J (2020). Efficient neuronal targeting and transfection using RVG and transferrin-conjugated liposomes. Brain Res 1734:146738.
- Bender HR, Kane S, Zabel MD (2016). Delivery of therapeutic siRNA to the CNS using cationic and anionic liposomes. J Vis Exp 113:e54106.
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, et al. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29(4):341-345.
- Xiang L, Zhou R, Fu A, Xu X, Huang Y, et al. (2011). Targeted delivery of large fusion protein into hippocampal neurons by systemic administration. J Drug Target 19(8):632-636.
- Kumar P, Wu H, McBride JL, Jung KE, Hee Kim M, et al. (2007). Transvascular delivery of small interfering RNA to the central nervous system. Nature 448(7149):39-43.
- Bu X, Yin C, Zhang X, Zhang A, Shao X, et al. (2019). LaSota strain expressing the rabies virus glycoprotein (rL-RVG) suppresses gastric cancer by inhibiting the alpha 7 nicotinic acetylcholine receptor (α7 nAChR)/Phosphoinositide 3-Kinase (PI3K)/AKT pathway. Med Sci Monit 25:5482.
- Balass M, Katchalski-Katzir E, Fuchs S (1997). The α-bungarotoxin binding site on the nicotinic acetylcholine receptor: Analysis using a phage-epitope library. Proc Natl Acad Sci USA (12):6054-6058.
- Javed H, Menon SA, Al-Mansoori KM, Al-Wandi A, Majbour NK, et al. (2016). Development of nonviral vectors targeting the brain as a therapeutic approach for Parkinson's disease and other brain disorders. Mol Ther 24(4):746-758.
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, et al. (2004). The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol 78(21):11487-11505.
- Järvelin AI, Noerenberg M, Davis I, Castello A (2016). The new (dis) order in RNA regulation. Cell Commun Signal 14:1-22.
- Norseen J, Johnson FB, Lieberman PM (2009). Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol 83(20):10336-10346.
- Raja MA, Katas H, Amjad MW (2019). Design, mechanism, delivery and therapeutics of canonical and Dicer-substrate siRNA. Asian J Pharm Sci 14(5):497-510.
- Dong-Ho K (2005). Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol 23:222-226.
- Amarzguioui M, Holen T, Babaie E, Prydz H (2003). Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res 31(2):589-595.
- Kim M, Shin D, Kim SI, Park M (2006). Inhibition of hepatitis C virus gene expression by small interfering RNAs using a tri-cistronic full-length viral replicon and a transient mouse model. Virus Res 122(1-2):1.
- Papayannopoulos V (2018). Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 18(2):134-147.
- Mackey D, Sugden B (1997). Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J Biol Chem 272(47):29873-29879.
- Glowinski J, Iversen L (1966). Regional studies of catecholamines in the rat brain-III: Subcellullar distribution of endogenous and exogenous catecholamines in various brain regions. Biochem Pharmacol (7):977-987.
- Lardizábal MN, Nocito AL, Daniele SM, Ornella LA, Palatnik JF, et al. (2012) Reference genes for real-time PCR quantification of microRNAs and messenger RNAs in rat models of hepatotoxicity. PloS one 7(5):e36323.
- Winsauer PJ, Daniel JM, Filipeanu CM, Leonard ST, Hulst JL, et al. (2011) Long‐term behavioral and pharmacodynamic effects of delta‐9‐tetrahydrocannabinol in female rats depend on ovarian hormone status. Addict Biol 16(1):64-81.
- Ibana JA, Belland RJ, Zea AH, Schust DJ, Nagamatsu T, et al. (2011) Inhibition of indoleamine 2, 3-dioxygenase activity by levo-1-methyl tryptophan blocks gamma interferon-induced Chlamydia trachomatis persistence in human epithelial cells. Infect Immun 79(11):4425-4437.
- Kopec AM, Rivera PD, Lacagnina MJ, Hanamsagar R, Bilbo SD (2017) Optimized solubilization of TRIzol-precipitated protein permits Western blotting analysis to maximize data available from brain tissue. J Neurosci Methods 280:64-76.
- Davarinejad H (2015) Quantifications of western blots with ImageJ. University of York.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi