Research Article, J Biochem Physiol Vol: 7 Issue: 2
Enhancement of Ca2+ Release from Store-Operated Ca2+ Entry by Corona Virus Disease 2019 (COVID-19) Spike (S) Protein
Kiwol Kim1, Eun Jeong Lee2 and Sang Sun Kang1*
11Department of Biology Education, Chungbuk National University, Chungdae-ro 1, Seowon-Gu, Cheongju, Chungbuk, 28644, Republic of Korea
2Department of Biochemistry Education, Chungbuk National University College Medicine, Chungdae-ro 1, Seowon-Gu, Cheongju, Chungbuk, 28644, Republic of Korea
*Corresponding Author: Kang SS,
Department of Biology Education, Chungbuk National University, Chundae-ro 1, Seowon-gu, Cheongju, Chungbuk, 361-763, Republic of Korea
Tel: +82432613278
E-mail: jin95324@cbu.ac.kr
Received date: 29 April, 2024, Manuscript No. JBPY-24-133442;
Editor Assigned date: 02 May, 2024, PreQc No. JBPY-24-133442 (PQ);
Reviewed date: 17 May, 2024, Qc No JBPY-24-133442;
Revised date: 25 May, 2024, Manuscript No JBPY-24-133442 (R);
Published date: 03 June, 2024, DOI: 10. 4172/jbpy.24.7.1000157
Citation: Kim K, et al. (2024) Enhancement of Ca2+ Release from Store-Operated Ca2+ Entry by Corona Virus Disease 2019 (COVID-19) Spike (S) Protein: Research Article. J Biochem Physiol 7:1.
Abstract
Functions and viral infection mechanisms of coronavirus disease 2019 (COVID-19) have been recently investigated extensively, focusing on Spike (S) protein together with its receptor, ACE2. Although their relationships with COVID-19 are obvious, less attention has been paid to intracellular regulation of S protein-protein interaction.
Here, we identified STIM1 (Stromal Interaction Molecule 1 Precursor) as a novel binding protein of S protein for the first time. Their association was further characterized. We found that S (1259DD1260) acidic motif specifically interacted with STIM1 C-terminal basic motif (671RKKFPLKIFKKPLKK685). Both motifs were demonstrated to be essential for STIM1 and S protein interaction using immune precipitation and immune blotting and confocal co-localization. We also elucidated that the association between the acidic tail motif of S protein and the C-terminal basic motif of STIM1 promoted Ca2+ cytoplasmic release from the Store- Operated Ca2+ Ion Entry (SOCE) by disrupting STIM1 function, suggesting that disrupting STIM1 function by S protein was one of its mode of actions for COVID-19 infection.
For the first time, we demonstrated that S protein played a role as a Ca2+ ion releasing enhancer for COVID-19 infection from SOCE through interrupting normal STIM1’s roles. Our findings may provide one of the new ways for curing or preventing COVID-19 pandemic recurrence.
Background: We noticed the presence of coronavirus disease 2019 (COVID-19) spike (S) protein acidic tail (1259DD1260) and STIM1 (Stromal Interaction Molecule 1 Precursor) C-terminal basic amino acid domain (671RKKFPLKIFKKPLKK685) by the consensus amino acid sequence survey.
Result: Association between S protein and STIM1 increases intracellular Ca2+ in the human cell, resulting in apoptosis and autophagy. Conclusion: Interaction between S protein and STIM1 causes the toxicity of COVID-19.
Significance: Binding between S protein and STIM1 that promotes the intracellular Ca2+ after its infection for the membrane fusion between host and COVID-19, assuming one of drug developing target points.
Keywords: COVID-19; Corona Virus; Spike Protein; Ca2+; Infection;
Immune; Blotting.
Introduction
After the outbreak of Coronavirus Disease 2019 (COVID-19) in late December 2019, it has brought serious harm and challenges to over 200 countries and regions around the world [1-3]. There is increasing evidence that many patients with COVID-19 are asymptomatic or they only have mild symptoms [4,5]. However, they can transmit the virus to others [6,7]. Although the pandemic has become an endemic with tremendous human wisdoms and efforts, COVID-19 contagion still has many dangerous aspects [8,9]. There are difficulties in screening for asymptomatic infections, which makes national prevention and control of this epidemic difficult [1-9]. However, human being should continue make efforts to research COVID-19 etiology.
Entry of coronaviruses into host cells is mediated by its spike glycoprotein (S protein). The Trans-membrane S protein can form Homo-trimers that starch from the viral surface. S protein is critical for the entry of coronaviruses [10-12]. Thus, it is an attractive antiviral target. It is composed of two functional subunits, S1 and S2. The S1 subunit consists of N-Terminal Domain (NTD) and Receptor Binding Domain (RBD). The function of S1 subunit is to bind to the receptor on host cell. S2 subunit contains Fusion Peptide (FP), Heptad Repeat 1 (HR1), Central Helix (CH), Connector Domain (CD), Heptad Repeat 2 (HR2), Trans-Membrane Domain (TM), and Cytoplasmic Tail (CT). The function of S2 subunit is to fuse the membranes of viruses and host cells [13]. The cleavage site at the border between S1 (1-685aa) and S2 (686-1273aa) subunits is called S1/S2 protease cleavage site [10-13] (Figure 1).
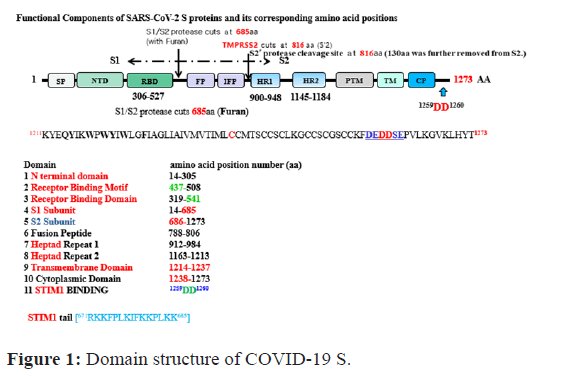
Figure 1: Domain structure of COVID-19 S.
The top and bottom numbers in each domain pertain to COVID-19 S, respectively. The red arrows indicate cleavage sites, and their numbers pertain to; Alignment of SP between COVID-19 S (top) and SARS-2-S (bottom); STIM1 (1257DEDDSE1262) has been reported to be the sensor of Ca2+ accumulation in the ER that activates store-operated Ca2+ entry. COVID 19 S protein contains conserved typical STIM1 domain-binding motif (1257DEDDSE1262) in its C-terminus (671RKKFPLKIFKKPLKK685), we speculate that the motif is also able to bind with STIM1. The modular protein- interaction domains and play important roles in receptor signaling and functions for the store-operated Ca2+ entry (SOCE). However, S mutant (1257DEAASE1262) does not interact with STIM1 C-terminus (671RKKFPLKIFKKPLKK685) and has no signal to SOCE. Alignment of two inter-domain segments; (E) COVID-19 S, (1257DEDDSE1262), COVID-19 S WT together with the top view of a helix showing hydrophobic positions a and d on the same side. The position of (1257DEDDSE1262) is indicated. Host proteases (eg. TMPRSS2) cleave the spike glycoprotein at the S2’ cleavage site to activate the proteins, which is critical to fuse the membranes of viruses and host cells through irreversible conformational changes generating S1(1-655aa), and S2’ (817-1273aa).
For all Coronaviruses, host proteases cleave the spike glycoprotein at the S2’ cleavage site to activate proteins S2’ (e.g., TMPRSS2) protease cleavage site at 816aa removing polypeptide (~130 aa) fragment. Such activation is critical for fusing the membranes of viruses and host cells through irreversible conformational changes generating S1 (1-655 aa) and S2’ (817-1273 aa).
Individual protein domains in the S protein tend to fold independently and are associated with specific functions [14,15]. The S protein contributes to host specificity and tissue specificity through its differential requirement of tissue-specific proteases [10-12]. The TM domain consists of the following four parts: A juxtamembrane aromatic part, a central hydrophobic part, a cysteine-rich part, and a highly hydrophilic cytoplasmic tail which anchors the spike inside the viral membrane together with the C-terminal acidic motif (1257DEDDSE1262) which attracts our attention the most [10-13]. Most studies on S protein have focused on its molecular mechanisms and physiological functions. However, much less attention has been given to intracellular regulation of S protein signaling, protein-protein interaction, and its cellular target point [10-15].
STIM1 consists of a 685-amino acid type I transmembrane protein originally identified in the plasma membrane, where it is oriented with its N-terminal domain targeting the extracellular space [16]. STIM1 was initially described as a protein that could induce growth arrest and degeneration of several Human Tumor Cell Lines, and was involved in stromal-hematopoietic cell interactions [17]. STIM1 has also been reported as a sensor of the amount of Ca2+ accumulated in the ER that activates Store-Operated Ca2+ Entry (SOCE), a mechanism for Ca2+ influx controlled by the filling state of intracellular Ca2+ stores [18,19]. When the intraluminal Ca2+ concentration is reduced, STIM1 (671RKKFPLKIFKKPLKK685) re-localizes within the ER membrane to punctae at ER-plasma membrane junctions, which facilitates its association with members of the Orai-1 and Transient Receptor Potential Cation (TRPC) families. STIM1 is able to gate TRPC1 through electrostatic interaction between the two conserved and negatively charged aspartates in TRPC1 (D639, D640) and the positively charged lysine residues in the STIM1 (671RKKFPLKIFKKPLKK685) motif located in the C-terminal polybasic region, as same as Orai-1 [20,21]. This motif can also bind to E cadherin (900DD901) C-terminal [22]. Through this process, STIM1 can activate SOC channels until ER Ca2+ levels are replenished [16-21].
In the course of our research on ACE2 (Angiotensin Converting Enzyme 2; known as COVID-19 S protein cell membrane receptor protein) tail motif (qTsF805) interaction with PSD95 PDZ III domain, we recognized the COVID-19 S protein C-terminal acidic motif (1259DD1260) which was assumed to be essential for its interaction with STIM1 (671RKKFPLKIFKKPLKK685) motif [1,22-25]. We have also reported that ACE2 containing a (799DD800) motif can interact with STIM1 through its basic motif [23,24]. These theoretical information led us again to pursue biological significances hiding in the association between COVID-19 S protein acidic tail (S1257DEDDSE1262) and STIM1 C-terminal basic amino acid motif (671RKKFPLKIFKKPLKK685) [1,22-25].
Consequently, we not only identified STIM1 as a protein associated with COVID-19 S protein, but also found that it could regulate basolateral localization of COVID-19 S protein via direct interaction. In this paper, we present experimental evidences for the protein-protein interaction between COVID-19 S protein and STIM1 and how COVID-19 S protein could disrupt STIM1 function through various mechanism such as Orai-1 interaction and SOCE [17-22]. Moreover, COVID-19 infection to host cells can trigger Ca2+ release from its reservoir through interaction of S protein with STIM1 to induce cell apoptosis. Thus, our findings provide not only one of the new ways to cure or prevent COVID-19 pandemic recurrence, but also a crucial clue to elucidate how the interaction contributes to COVID-19 virulence [10-16].
Materials and Methods
Preparation of plasmids
COVID-19 S was purchased from GeneCopia (EX-COV219-M98). Spike protein (1259AA1260) motif mutant was constructed with primers (Up: 5’-AAA TTT GAT GAA GCC GCC TCT GAG CCA GTG-3’ Down: 5’-CAC TGG CTC AGA GGC GGC TTC ATC AAA TTT-3’) in PCR. pGW-Cherry-STIM1 constructs were kindly provided by Dr Jong Guk Choi (Chungbuk National University, Cheungju Korea). All of the DNA constructs were individually verified by DNA sequencing.
Western blotting and antibodies
Samples were run on SDS/PAGE and then transferred on to a nitrocellulose membrane. The membrane was blocked in TBST (TBS (20 mM Tris/HCl, pH 7.5, and 500 mM NaCl) with 0.05% Tween 20) containing 5% (w/v) non-fat dried milk for 1 h at room temperature (23°C-25°C). Proteins of interest were probed with corresponding primary antibodies followed by HRP (Horseradish Peroxidase) conjugated anti-rabbit or anti-mouse secondary antibodies. Immunoreactive bands were visualized by ECL detection reagents (Applygen Technologies) and analyzed with NIH Image 1.62.
Anti-EGFP antibody was from Applygen Technologies. Anti- STIM1 antibody was obtained from Sigma-Aldrich. Anti-COVID 19 S protein antibody and S1 protein was from Novus Biologicals. Anti-S protein was from Santa Cruz Biotechnology.
Cells and transfection
HEK293 (Human Embryonic Kidney 293) and NIH 3T3 cells were maintained in DMEM (Dulbecco’s Modified Eagle’s Medium) plus 10% (v/v) FBS and 1% Penicillin/StreptoCherryin (Sigma-Aldrich). The cells were cultured on polylysine-coated glass coverslips in DMEM supplemented with 10% (v/v) FBS, 10% (v/v) Horse Serum and 1% Penicillin/StreptoCherryin. Cell transfection was performed with Lipofectamine™ 2000 (Invitrogen). For the stable cell line, constructs of GFP; COVID 19 S protein WT or GFP; COVID 19 S (1259AA1260) mutant were transfected into HEK293 or NIH 3T3 cells respectively with Lipofectamine™ 2000, and selected with the growth medium containing 1200 μg/ml neomycin G418 (Amresco) for a month. We also established the transient cell line which is expressing COVID-19 WT S protein or S (1259AA1260) mutant from HEK293 and NIH 3T3 for the experiment purpose.
Co-Immunoprecipitation
Co-Immunoprecipitation was performed as described previously. Briefly, transfected cells were harvested or rabbit brain was homogenized in ice-cold lysis buffer. Supernatants were incubated with anti-EGFP beads (for transfected cells) or anti-STIM1 antibody (for brain lysates) pre-bound to Protein A/G–agarose beads (Calbiochem). The Immunoprecipitated proteins were then analyzed by western blotting.
Immunnofluorescence co-localization
Co-localization analysis with immunofluorescence was performed as described previously [21]. Briefly, HEK293 cells were transfected with GFP COVID-19 S protein and/or STIM1 constructs. After fixation and permeabilization, cells were stained with anti-STIM1 antibody and anti-S antibody (Abcam) followed by rhodamine- conjugated anti-(mouse IgG) antibody (ZSGB-BIO), Alexa Fluor® 647-conjugated anti-(rabbit IgG) antibody (Invitrogen) and Hoechst 33258 (5 μg/ml). The cellular distribution of these proteins was then visualized under a confocal microscope (Leica Microsystems, LAS AF-TCS SP5).
Fluorescence measurements of (Ca2+)i
We measured (Ca2+)i using a fluorescent Ca2+ indicator Fluo4- acetoxymethyl ester (Fluo4-AM), as previously described (24-26). In brief, the stable (or transiently) cells transfected with EGFP–COVID 19 S protein WT or EGFP–COVID 19 S (1259AA1260) mutant growing on coverslips were incubated for 40 min in DMSO solution containing 1 μM Fluo4-AM at 24°C in darkness, and then washed and incubated for 15 min to hydrolyze internalized Fluo4-AM. We measured (Ca2+) i in single cells that emitted fluorescence, using confocal microscopy (LSM710 Zeiss, Germany) at wavelengths of 495 nm (excitation), and 519 nm (emission). The absorption (as an arbitrary unit) at 488 nm with an argon-ion laser was measured as a relative intracellular Ca2+ ion concentration (Ca2+)i. All experiments were carried out at 24°C.
FACS (Fluorescence-Activated Cell Sorting)
FACS was performed with our previous method in elsewhere. COVID-19 Spike, its AA mutant and EGFP vector, were transfected into cells and the rates of apoptosis measured with an Annexin V-PE apoptosis detection kit I (BD Biosciences, USA). Transfected cells were washed twice in cold PBS and resuspended in binding buffer [0.01 M Hepes/NaOH (pH 7.4) 0.14 M NaCl, 2.5 mM CaCl2]. 1 × 105 cells in 100 ml were transferred to 5 ml culture tubes and 5 ml of Annexin V-PE and 5 ml of 7-Amino-actinomycin D were added. The cells were vortexed and incubated for 15 min at 25°C in the dark. 400 ml of binding buffer was added to each tube. Within 1 h FACS was performed on a Coulter Epics Elite equipped with a gated amplifier and upgraded to give enhanced system performance in the Core Facility of Chungbuk National University.
Results
STIM1 (671RKKFPLKIFKKPLKK685) is identified as a novel COVID-19 S protein acidic motif-binding protein (1257DEDDSE1262)
With the previous many biological data base information that STIM1 (671RKKFPLKIFKKPLKK685) binds with the acidic motif (DD), we recognized that COVID 19 S protein contains (1257DEDDSE1262) motif in cytoplasmic tail resign (Figure 1) [1-16].
This information led us to study the S protein-STIM1 interaction and its biological significance behind this interaction. The amino acid number on Figure 1 was adapted from the coronavirus which is published at the first [1].
S protein is divided two functional subunits, S1 and S2. The S1 subunit consists of N-Terminal Domain (NTD) and Receptor Binding Domain (RBD). The function of S1 subunit is to bind to the receptor on host cell. S2 subunit contains Fusion Peptide (FP), Heptad Repeat 1 (HR1), Central Helix (CH), Connector Domain (CD), Heptad Repeat 2 (HR2), Transmembrane Domain (TM), and Cytoplasmic Tail (CT).
The function of S2 subunit is to fuse the membranes of viruses and host cells. The cleavage site at the border between S1 (1-685aa) and S2 (686-1273aa) subunits is called S1/S2 protease cleavage site and furan [10-13].
The host proteases cleave the spike glycoprotein at the S2’ cleavage site to activate proteins S2’ (e.g., TMPRSS2) protease cleavage site at 816aa of S2, removing polypeptide (~130 aa) fragment. The cleavage is known as the critical step for fusing the membranes of viruses and host cells through irreversible conformational changes generating S1 (1-655 aa) and S2’ (817-1273 aa) [11-13].
STIM1 (671RKKFPLKIFKKPLKK685) also interacts with COVID-19 S protein (1257DEDDSE1262)
With the information that STIM1 poly basic amino acid tail prefers to bind with the short acidic motif (DD) of its partner protein, we noticed the motif (1257DEDDSE1262) on S protein.
At first, we determined whether the protein-protein interaction between STIM1 and S protein occurred. For this purpose, we generated COVID-19 S protein expression cell line from HIH3T3 cell or HEK293 cell line with COVID-19 S protein expression vector which was purchased from GenCopia Co. We constructed COVID 19 S protein expression cell lines from NIH/3T3 cell or HEK293 background and observed the same protein-protein complexes between S protein and STIM1 in Hela cells and Human Liver Tissue Lysates (data not shown). Next, we tested whether S protein interacts with STIM1 using IB and IP tool. We conformed that COVID-19 S protein interacts with STIM1 in HEK293 by the immunological method (Figure 2).
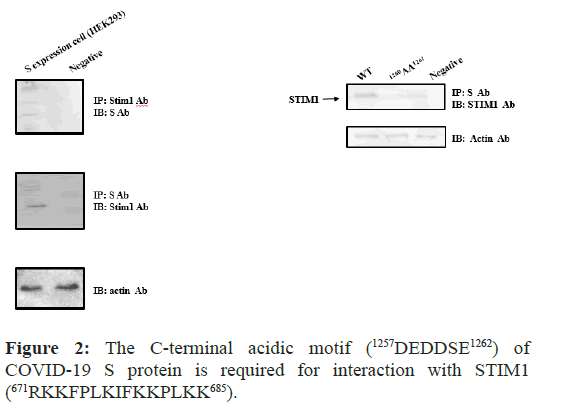
Figure 2: The C-terminal acidic motif (1257DEDDSE1262) of COVID-19 S protein is required for interaction with STIM1 (671RKKFPLKIFKKPLKK685).
With S protein expression stable cell line or transient transfected cell, the protein-protein interaction between Spike and STIM1 in HEK 293 cells was performed. Following Immunoprecipitation (IP) using an anti-ACE2 antibody, Immunoblot (IB) analysis was performed using an antibody against STIM1 (left). STIM1 Immunoprecipitated complexes were subjected to Immunoblot analysis using an anti-S antibody (right) Co-immunoprecipitation of STIM1 with S protein confirmed the formation of a S protein–STIM1 complex. The negative control for immunoprecipitation was an unrelated antibody. The control for western blot analysis was an antibody against actin (bottom).
To define the binding sites, both (1259DD1260) of S protein and of STIM1, we generated the mutants with the site directed mutagenesis (1259AA126). With the mutant, we also performed IP and IB experiments in (Figure 2).
As shown in Figure 2 the mutant did not interact with STIM1, while the WT (1259DD1260) did pull down STIM1 Therefore, for the first time, we demonstrated that COVID 19 S protein interacts with STIM1. The interaction requires S protein (1259DD1260) motif, as we expected (Figure 1).
COVID-19 S protein (1259DD1260) and STIM1 association contributes to the subcellular localization of COVID 19 S protein.
HEK293 cells were transiently transfected with GFP-COVID-19 S protein WT, or GFP–COVID-19 S protein (1259AA1260), together with Cherry–STIM1. Co-Localization of COVID-19 S protein (1259DD1260) with STIM1 (671RKKFPLKIFKKPLKK685) was shown in the yellow merging of the two individual images, as shown in panels (Figure 3). The results also support the protein-protein interaction between S protein and STIM1 by the co-localization of COVID-19 S protein with STIM1 at Golgi, together with IP and IB experiments as shown in Figure 2.
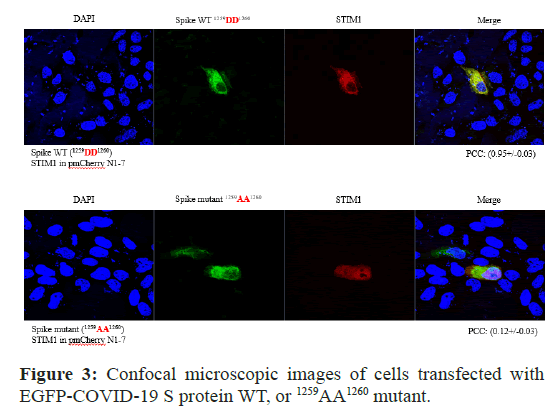
Figure 3: Confocal microscopic images of cells transfected with EGFP-COVID-19 S protein WT, or 1259AA1260 mutant.
A Cells were examined by direct immunofluorescence microscopy. The figures show EGFP-COVID-19 S protein WT or (1257DEAASE1260) mutant (green), STIM1 (red), and merged (yellow) confocal microscopic images. EGFP-COVID-19 S protein WT or 1257DEAASE1260 showed minimal co-localization with STIM1 at the Golgi apparatus (B left and right). EGFP-COVID-19 S protein or (1259AA1260) was principally detected in the plasma membrane (right). However, EGFP-COVID-19 S protein WT was co-localized with STIM1 at the Golgi apparatus (B middle). Fn/c (factional rate of nuclear localization to cytoplasm and PCC (Pearson Co-localization co-efficiency) are indicated below.
However, as shown in Figure 3, GFP–COVID-19 S protein (1259AA1260) did not merge with Cherry–STIM1 (its PCC was (0.12+/- 0.03). Thus, S protein (1259DD1260) motif is required to interact and co-localize with STIM1 in the cell together at Golgi (Figure 3). The PCC was (0.95+/-0.03). Together, those results also confirmed that COVID-19 S protein (1259DD1260) motif interacts with STIM1.
Ca2+ live image comparison of COVID-19 S WT, or mutant (1259AA1260) in HEK cells
To compare with the protein-protein interaction on the effect of intracellular calcium concentration, the confocal microscopic Ca2+ live images of the transfected EGFP-COVID-19 S, WT, or (1259AA1260) mutant were conducted. The image is representative of five repeat experiments. Effects of S protein treatment on intracellular calcium concentration change (Ca2+)i of COVID-19 S protein WT, or (1259AA1260) mutant was measured by the absorption at 488 nm of argon-ion laser in HEK 293 cells (as an arbitrary % unit .
As shown in Figure 4, COVID-19 S protein WT (A) enhanced (Ca2+)i in HEK293 almost ten times higher than COVID-19 S (1259AA1260) mutant did (B). Conversely, the results suggested that the protein-protein interaction between COVID-19 S protein WT and STIM1 through (1259DD1260) motif contributes (Ca2+)i enhancement.
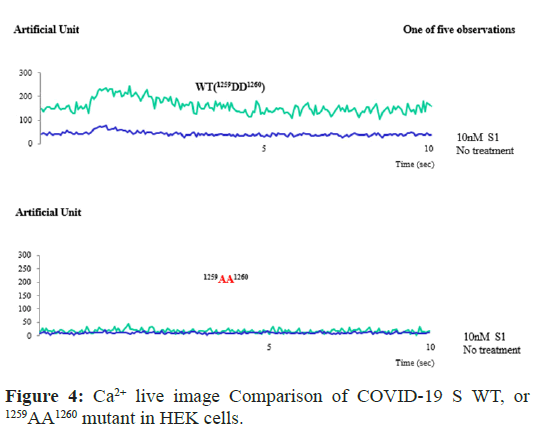
Figure 4: Ca2+ live image Comparison of COVID-19 S WT, or 1259AA1260 mutant in HEK cells.
Confocal microscopic analysis of the transfected EGFP- COVID-19 S protein WT, or 1259AA1260 mutant (green). The localization of COVID-19 S protein WT was altered by treatment with IGF1. The image is representative of five repeat experiments. Effects of heat on intracellular calcium concentration change (Ca2+)i of COVID-19 S protein WT, or 1259AA1260, expressed by the absorption at 488 nm of argon-ion laser in HEK 293 cells (as an arbitrary % unit).
The abnormal regulation of STIM1 by S protein in the cytosol Ca2+ release after COVID-19 infection
In case of Spike (S2’) protein association with STIM1, S2’protein cytoplasmic (1259DD1260) motif can bind with STIM1 (671RKKFPLKIFKKPLKK685) and stimulate to form STIM1 channel complexes which transport Ca2+ in the cytoplasm from ER, resulting in Ca2+ release in the cytoplasm from ER. The release Ca2+ in the cytoplasm seems to stimulate the cell apoptosis or further the COVID-19 infection through the stimulation membrane fusion between plasma membrane and viral membrane. In infection state (Figure 5)
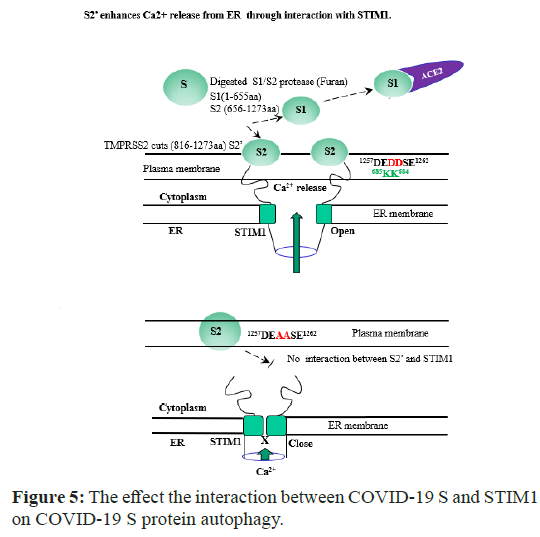
Figure 5: The effect the interaction between COVID-19 S and STIM1 on COVID-19 S protein autophagy.
In COVID-19 S transfect state, the host S1/S2 protease with furan cleavage S protein between 655-656 aa peptide bond, generating S1(1- 655aa) and S2 (656-1273aa). S1 protein still binds with its host cell ACE2 receptor. However, S2 subunit is processed further by the other host proteases (eg.TMPRSS2). For all the coronaviruses, TMPRSS2 cleaves at the S2 specific site (815-816 aa) to generate S2’ (816- 1273aa) which is critical to fuse the membranes of viruses and host cells through irreversible conformational changes. S2’ (816-1273aa) exposes the C-terminal acidic motif (1257DEDDSE1262) forward the basic motif tail (671RKKFPLKIFKKPLKK685) of STIM1 in ER. This event stimulates SOCE to release Ca2+ in the cytoplasm. The increased Ca2+ in the cytoplasm seems to promote the cell apoptosis (Table 1). In our S protein expression cell line, S protein was also followed the similar process (Figure 2). In the contrast, S (1257DEAASE1262) mutant cannot bind to STIM1 (671RKKFPLKIFKKPLKK685) nor form STIM1 channel complex. Thus, the transport Ca2+ in the cytoplasm from SOCE is blockaded (Figure 4), and less cytotoxic effect.
Table 1: Comparison cell survival between COVID19 S WT and 1259AA1260 mutant. COVID-19 S contain acidic tail motif 1259DD1260 which interacts with STIM1 basic tail. We compared S WT protein cytotoxicity (or apoptosis) with 1259AA1260 mutant in FACS. The results showed that S interaction with STIM1 effects on host cell survival with Ca2+ release in the cytoplasm.
However, S2’ protein (1259AA1260) mutant is not able to bind with STIM1 (671RKKFPLKIFKKPLKK685) nor form STIM1 channel complex. Thus, the transport Ca2+ in the cytoplasm from ER is blockaded (Figure 5). As shown in Figure 4 and Table 1, we clearly observed that the protein-protein interaction causes cell autophagy and apoptosis in both NIH3T3 and HEK293 cell. This observation also suggests that the disruption of STIM1 interaction by COVID-19 S protein motif (1259DD1260) causes Ca2+ leak from Endoplasmic Reticulum (ER) or extracellular Ca2+ reservoir to cytoplasm, resulting in cell apoptosis (Table 1).
In summary, S protein cytoplasmic (1259DD1260) motif is required to interact with STIM1. The association between S protein and STIM1 enhances the intracellular Ca2+ concentration which contributes host cell apoptosis and cytotoxicity.
Discussion
We attempted here to demonstrate this topic whether COVID-19 S protein binds to STIM1 and how the interaction contributes to disrupts its signaling and function, since the modular protein- interaction domains play important roles in receptor signaling and functions (Figure 1 and Figure 5).
With the positive-charged lysine residues in STIM1’s C-terminal polybasic region (671RKKFPLKIFKKPLKK685), STIM1 is known to play an important role in the trafficking of transmembrane proteins, such as Orai-1, TRPC1, ACE2 (Angiotensin Converting Enzyme 2), E-cadherin, and TRPV4 [16-22]. After recognizing that COVID-19 S protein contains a conserved typical STIM1 domain binding motif (1257DEDDSE1262) in its C-terminus, we speculated that the motif is also able to bind with STIM1 [1,16-22].
Starting from the line of this speculation, we characterized here that STIM1 (671RKKFPLKIFKKPLKK685) is also a novel COVID-19 S protein-interacting molecule in the cytosol. The C-terminus of COVID-19 S protein (1257DEDDSE1262) motif specifically interacts with the motif of STIM1. In our present study of COVID-19 S protein functions, for the first time we found that the motif (1257DEDDSE1262) in S protein C-terminal interacts with STIM1 (Figure 5).
Consequently, we demonstrated here that COVID-19 S protein contains a conserved typical STIM1 domain-binding motif in its CT (C-terminus) (Figure 1). The motif is able to bind to STIM1 domains which are modular protein-interaction domains and play important roles in receptor signaling and functions (Figure 5). Also, we identified that STIM1 associates with the COVID-19 S protein and the stabilization of the COVID-19 S protein is regulated by STIM1 via direct interaction and possibly through its apical membrane localization and the inhibition of COVID-19 S protein (Figure 3). Investigating the COVID-19 S protein-binding molecules and its post-translational modifications are therefore of great interest to combat with COVID-19.
Functional studies showed that COVID-19 S protein contains its acidic tail (1257DEDDSE1262) motif to promote Ca2+ release through binding with STIM1 basic amino acid tail motif (671RKKFPLKIFKKPLKK685). Therefore, our findings reveal a new mechanism for the regulation of STIM1 by COVID-19 S, involving the inhibition of STIM1 channel function. The elucidation of COVID-19 S protein-mediated regulation of Ca2+ concentration mechanism may help to clarify further the functions of the COVID-19 S protein in the Renin-Angiotensin System (RAS) (Figure 5).
The plasma membrane protein of these coronaviruses recognizes the receptors such as ACE2 and undergoes cell fusion and S protein needs to be cleaved by the host protein before fusion with the membrane. This process is necessary for the virus to infect the host and blocking this process can prevent viral infection [10-13].
Here, we identified STIM1 (Stromal Interaction Molecule 1 Precursor) as a novel binding protein of COVID 19 S protein (1257DEDDSE1262), and their protein association requires to be characterized further. S protein specifically interacts with STIM1 C-terminal basic motif (671RKKFPLKIFKKPLKK685). With GST pull-down, co-immunoprecipitation, and confocal co-localization experiments, the interaction between S protein and STIM1 was proven. Further, gain-of-function and loss-of-function studies indicated that S protein motif interaction with STIM1 enhances cytoplasmic Ca2+ concentration from ER, resulting in cell apoptosis. The detail mechanism how the enhanced cytoplasmic Ca2+ concentration causes the cell apoptosis remains to be elucidated in the future. COVID-19 infection to the host cell triggers Ca2+ ion release from its reservoir through promoting the interaction of ACE2 protein with STIM1.
Because STIM1 also seems to play an important role in the trafficking or subcellular localization of transmembrane proteins such as Orai-1, TRPC1, and E-cadherin, S protein motif interaction with STIM1 may disturb the proper localization of these proteins [16-22]. Moreover, as ACE2 is also known as a Zn2+ binding protein for its protease activity, it seems to function as a Zn2+ or other ion trafficker protein from the extracellular milieu into the endoplasmic reticulum, similar to Orai-1 for Ca2+. Further studies are necessary to confirm this hypothesis, and we pursue this possibility [23-25].
The interaction between S protein acidic motif (1257DEDDSE1262) and STIM1 C-terminal basic motif (671RKKFPLKIFKKPLKK685) may also contribute to the COVID-19 cytotoxicity through Ca2+ cytoplasmic release from the store-operated Ca2+ ion entry through disrupting STIM1 function (Figure 5). Thus, the increased interaction between the acidic motif (1259DD1260) in S protein and STIM1 through COVID-19 S may inhibit the overall STIM1 functions by the disruption of its normal protein-protein interaction or subcellular localization, resulting in COVID-19 virulence [14-25]. However, the other mode of actions for COVID-19 virulence by S protein remains to be still elucidated [1-9]. ACE2 is well characterized in its role in RAS which is the important role to maintain the individual physiological normal balance [10-13]. Because of this reason, COVID-19 may also evolve to adapt the host physiology for maximizing its survival rate, as the nature anti-reaction force for the equilibrium. Thus, ACE2 seems to be naturally selected as the effective COVID-19 receptor during the virus evolution. After S protein accumulation in the host cell, it also contributes to the dissociation of ACE2 from STIM1 with its (1257DEDDSE1262) motif as a feedback inhibition/feed forward activation for the viral survival (Figure 5). The mode of action of S protein to its host cell seems to be similar to that of COVID 19 E protein which contains PDZ motif, completive to PDZ domain interaction [23-25].
From this assumption, we also speculated that the C-terminus of COVID-19 S protein (1257DEDDSE1262) motif which specifically interacts with the motif of STIM1 seems to be evolved for the viral maximum survival purpose through COVID-19 natural selection process. Because STIM1 is the most essential and multifunctional point for host cell survival regulation (such as Ca2+ on regulation), the host target (disruption) motif of COVID-19 S protein interaction seems to be selected for its maximum survival effectiveness. After COVID-19 infection, the S protein also localizes to the host cell basolateral membrane by STIM1 binding.
There is still a long way to go in research efforts to combat the virus. We hope that this struggle between humans and microorganisms can permit us to understand our viral opponents more carefully. For the first time, we demonstrated that COVID-19 S protein also functions as a host Ca2+ ion influx modulator protein for its infection and toxicity (Figure 5).
Conclusion
In conclusion, here for the first time, we identified that STIM1 is a COVID-19 S association protein through its tail acidic motif (1257DEDDSE1262) and the subcellular of the COVID-19 S protein is regulated by STIM1 via direct interaction and possibly through its basolateral membrane localization and the inhibition of STIM1.
Furthermore, COVID-19 infection to the host cell triggers Ca2+ release from its reservoir through the disruption of the interaction of Orai1 with STIM1 by S protein, inducing cell autophagy and apoptosis. Even though most studies on S protein have focused on its molecular mechanisms and physiological functions, but much less attention has been given to the intracellular regulation of S protein signaling, protein-protein interaction, and its cellular target point.
References
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265-269.
[Crossref] [Google Scholar] [PubMed]
- Baker RE, Mahmud AS, Miller IF, Rajeev M, Rasambainarivo F, et al. (2022) Infectious disease in an era of global change. Nat Rev Microbiol. 20(4):193-205.
[Crossref] [Google Scholar] [PubMed]
- Chauhan S. (2020) Comprehensive review of Coronavirus Disease 2019 (COVID-19).Biomed J.43(4):334-340.
[Crossref] [Google Scholar] [PubMed]
- Colaneri M, Novelli V, Cutti S, Muzzi A, Resani G, et al. (2021) The experience of the health care workers of a severely hit SARS-CoV-2 referral Hospital in Italy: Incidence, clinical course and modifiable risk factors for COVID-19 infection. J Public Health 43(1):26-34.
[Crossref] [Google Scholar] [PubMed]
- Hernández-Torres R, MartÃnez Lozano M, Torres I, Rosario-Hernández E, Ramos-Pibernus A, et al. (2023) The impact of the COVID-19 pandemic and previous natural disasters on the mental health of healthcare workers in Puerto Rico. PLOS Glob Public Health.17;3(5):e0001784.
[Crossref] [Google Scholar] [PubMed]
- Jiménez Padilla EA, Galindo Vázquez O, Jiménez Flores J, Costas Muñiz R, Meneses GarcÃa A. (2023) Factors associated with the presence of fear of contagion and burnout syndrome in nursing personnel during the COVID-19 pandemic. J Res Nurs. 28(6-7):418-431.
[Crossref] [Google Scholar] [PubMed]
- Adedokun KA. (2021) Early stage nonclinical pulmonary disorder in COVID-19 may present asymptomatic and fuel the contagion. Mil Med Res. 8(1):22.
[Crossref] [Google Scholar] [PubMed]
- Chung HW, Apio C, Goo T, Heo G, Han K, et al. (2021) Effects of government policies on the spread of COVID-19 worldwide. Sci Rep 11(1):20495.
[Crossref] [Google Scholar] [PubMed]
- Choi H, Lee JH, Park HK, Lee E, Kim MS et al. (2022) Impact of the COVID-19 pandemic on patient delay and clinical outcomes for patients with acute myocardial infarction. J Korean Med Sci. 37(21).
[Crossref] [Google Scholar] [PubMed]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181(2):271-280.
[Crossref] [Google Scholar] [PubMed]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, et al.(2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 117(21):11727-11734.
[Crossref] [Google Scholar] [PubMed]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, et al. (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 81(2):281-292.
[Crossref] [Google Scholar] [PubMed]
- Wang Q, Zhang Y, Wu L, Niu S, Song C, et al. (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 181(4):894-904.
[Crossref] [Google Scholar] [PubMed]
- Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, et al. (2020) The molecular virology of coronaviruses. J Biol Chem. 295(37):12910-12934.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Zhao X, Zhou H, Zhu H, Jiang S, et al. (2023) Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat Rev Immunol. 23(3):189-199.
[Crossref] [Google Scholar] [PubMed]
- Oritani K, Kincade PW. (1996) Identification of stromal cell products that interact with pre-B cells. J Cell Biol. 134(3):771-782.
[Crossref] [Google Scholar] [PubMed]
- Sabbioni S, Veronese A, Trubia M, Taramelli R, Barbanti-Brodano G, et al. (1999) Exon structure and promoter identification of STIM1 (alias GOK), a human gene causing growth arrest of the human tumor cell lines G401 and RD. Cell Genet. 86(3-4):214-218.
[Crossref] [Google Scholar] [PubMed]
- Luo R, Le Gourriérec P, Antigny F, Bedouet K, Domenichini S, et al. (2024) STIM2 variants regulate Orai1/TRPC1/TRPC4-mediated store-operated Ca2+ entry and mitochondrial Ca2+ homeostasis in cardiomyocytes. Cell Calcium.119:102871.
[Crossref] [Google Scholar] [PubMed]
- Feske S. (2007) Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 7(9):690-702.
[Crossref] [Google Scholar] [PubMed]
- Feske S, Prakriya M, Rao A, Lewis RS. (2005) A severe defect in CRAC Ca2+ channel activation and altered K+ channel gating in T cells from immunodeficient patients. J Exp Med. 202(5):651-662.
[Crossref] [Google Scholar] [PubMed]
- Huang GN, Zeng W, Kim JY, Yuan JP, Han L et al. (2006) STIM1 carboxyl-terminus activates native SOC, I crac and TRPC1 channels. Nat Cell Biol. 8(9), 1003-10010.
[Crossref] [Google Scholar] [PubMed]
- Hong JH, Li Q, Kim MS, Shin DM, Feske S, et al. (2011) Polarized but differential localization and recruitment of STIM1, Orai1 and TRPC channels in secretory cells. Traffic 12(2)232-245.
[Crossref] [Google Scholar] [PubMed]
- Lee EJ, Shin S, Hyun S, Kang SS. (2021) ACE2 tail motif (QTSF-COOH805) regulates ACE2 apical membrane localization and cell growth. Baltica J. 34(3):15-44.
- Lee EJ, Kim K, Davaadorj O, Shin SH, Kang SS. (2024) Assessment Interaction between PSD 95 and TRPV4 through PDZ Domain Controls TRPV4â??s Localization and Activity. J Anal Tech Res. 6:7-18.
- Lee EJ, Shin SH, Hyun S, Kang SS. (2021) COVID-19 envelope (E) plays an antagonistic role against ACE2 through its PDZ tail motif. Polish Polar Research. 42(3):52-67.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi