Research Article, Vol: 12 Issue: 3
Does Protein Kinases exhibit Binding Affinity with Antipsychotic Drugs? – A Molecular Docking Approach
Kiran.P.C1, Sarojini.R.Bulbule1, Ramesha Hanumanthappa1, Hemalatha Nanjaiah2, and Devaraju K S1*
1Department of Biochemistry, Karnatak University, Dharwad, India
2Department of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, USA
*Corresponding Author: Devaraju K S,
Department of Biochemistry, Karnatak
University, Dharwad, India
Tel: +91-9164965420
E-mail: ksdevaraju@kud.ac.in
Received date: 21 June, 2023, Manuscript No.JABCB-23-103452;
Editor assigned date: 23 June, 2023, PreQC No. JABCB-23-103452 (PQ);
Reviewed date: 11 July, 2023, QC No. JABCB-23-103452;
Revised date: 18 July, 2023, Manuscript No. JABCB-23-103452 (R);
Published date: 24 July, 2023, DOI: 10.4172/2327-4360.1000266
Citation: Kiran PC, Bulbule SR, Hanumanthappa R, Nanjaiah H, Devaraju KS (2023) Does Protein Kinases exhibit Binding Affinity with Antipsychotic Drugs? – A Molecular Docking Approach. J Appl Bioinforma Comput Biol 12:3
Abstract
Introduction: Antipsychotic drugs are used to treat psychosis and majorly used are phenothiazine drugs like Clozapine, Olanzapine, and Trifluoperazine. Both typical and atypical antipsychotic drugs are known to bind serotonin, dopamine, histamine, muscarine receptors and other associated proteins. However, it is not known that these antipsychotic drugs would bind to kinases are not. Kinases have emerged as the largest family of signaling proteins in eukaryotic cells and are involved in every aspect of cellular regulation. All these antipsychotic drugs crossing plasma membrane and found to be in cytosol have all the possibility of interacting with the other molecules. On the other hand all antipsychotic drugs without an exception they exhibit side effects as Food and Drug Administration (FDA) approved drowsiness, dizziness, restlessness, weight gain so on.
Method: The current study is carried out to know whether these antipsychotic drugs bind to different various protein kinases or not, this was done by using Molecular docking analyses which were performed using PyMol, Discovery Studio Biovia 2017, AutoDock Vina, AutoDock Tools version 1.5.4 and LigPlot to evaluate their binding free energy (ΔG).
Results: Among all the compounds, Trifluoperazine exhibited the higher binding affinity of – 9.1 Kcal mol-1 with Casein kinase 1 when compared to Clozapine and Olanzapine.
Conclusion: Further his study suggests that the Trifluoperazine exhibits a non-specific modulatory effect on kinases in general, Casein kinase I in particular. This need to be further explored In Vitro and molecular level studies.
Keywords
Antipsychotics; Protein kinases; Binding affinity; Free energy; Auto dock
Abbreviations
PK: Protein Kinase; CLZ: Clozapine; OLZ: Olanzapine; AMPK: AMP –actuated Protein Kinase; TFZ: Trifluoperazine; KV: Voltagedependent K; Kcal mol-1: Kilo Calories per Mole; PDB: Protein Data Bank; SDF: Spatial Data File; PLP: Protein Ligand Filters
Introduction
Phosphorylation is a major component of protein synthesis and is involved in the control of enzyme activity. It is also fundamental to cell signaling. [1]. The role of phosphotransferases in cellular processes is widely acknowledged as a target for the development of effective drugs for the treatment of various diseases. Currently, around 540 Protein Kinase (PK) have been identified and are being studied in various phases of drug discovery [2]. Generally, numerous of inhibitors are available to study the binding ability with the targeted proteins [3]. A gene is a sequence of Deoxyribonucleic Acid (DNA) or Ribonucleic Acid (RNA) that enables the production of a protein or a gene product. Serine-threonine protein kinase, is a member of the CAMK subfamily, has been shown that it regulates the activity of various cellular genes [4]. These kinases are targeted for the next generation of notable antipsychotic drugs and many other too; Phenothiazines belong to the oldest, synthetic antipsychotic medicines, and don’t have their precursor in the world of natural composites. These medicines are fairly stable and extensively used as antipsychotic medicines that substantially act on central dopamine receptors and have opiate, antiemetic, anti-psychotic and body temperature-lowering goods [5].
Antipsychotic medicines are presently used in clinical practice for a variety of internal diseases. Among them, Clozapine (CLZ) is the most effective drug for treatment-resistant schizophrenia and is most helpful in controlling aggression and the suicidal geste in schizophrenia and schizoaffective complaint [6].The targets interrogated included CLZs validated target (eg: D2, D3 dopamine; 5-HT2A serotonin) as well as utmost of the presently delved targets (eg: M1, M4 muscarinic; mGluR2/3 metabotropic glutamate; nicotinic acetylcholine). The most likely locus of CLZ conduct was engagement of the serotonergic neuronal system because numerous serotonin receptors are both preand post-synaptic with respect to serotonin neurons. Indeed, former studies demonstrated that CLZ and affiliated atypical antipsychotic medicines have preferentially high for postsynaptic 5-HT2A serotonin, D4 dopamine, muscuraine, acetylcholine as well as dozens of GPCRs. Each of these individual molecular targets to which CLZ binds with affinity has been latterly exploited for medicine discovery purposes [7].
On the other hand Olanzapine (OLZ) is a atypical antipsychotic agent used for treatment of schizophrenia. It is associated with side effects of metabolic abnormalities, weight gain and impaired glucose tolerance. This is suggested to be mainly through autophagy subsequent to the activation of AMP-activated Protein Kinase (AMPK). Through activation of autophagy through the AMPK pathway, OLZ is suitable to meliorate the injury convinced by Rotenone, a neurotoxic germicide, in the PC 12 cell line. This has been delved that OLZ is implicit to inhibit Glycogen Synthase Kinase 3 (GSK3) due to their hypoglycemic side goods [8]. It’s also been observed that direct effect of OLZ on the hepatic insulin signaling pathway, with a reduction in Insulin Receptor Substrate 2 (IRS2) situations in the phosphorylation of GSK3a-Ser21, and an increase in the phosphorylation of GSK3b- Ser9, which appears in the absence of an effect on weight or visceral adipose towel deposit [9].
Trifluoperazine (TFZ) is a calmodulin asset and classic anxiolytic and antipsychotic medicine [10]. Former examinations have demonstrated that TFZ can arrest cell cycle [11], inhibit cell proliferation [12] and induce apoptosis [13]. Former studies have reported TFZ suppressed the proliferation of fibrosarcoma HT1080 leukemia, bone cancer and mortal A549 lung adenocarcinoma cells by regulating different signaling pathways [14]. In studies of phenothiazine medicine in which the dose-dependent effects of phenothiazines on the viability of leukemic cells and normal lymphocytes have been investigated in parallel, and demonstrating that the drugs (at clinically relevant doses) express selective cytotoxicity and antiproliferative activity against cancer cells without any influence on the viability of normal lymphocytes [15].
There are several reports of TFZ, in its capacity as a CaMK II asset, modulating ion channels in a CaMK II-independent manner. TFZ inhibited voltage-dependent K (Kv) channels in rabbit coronary arterial smooth muscle cells and Kv 1.3 channels expressed in mortal T lymphocytes anyhow of its function as a calmodulin assest [16]. It also blocks both Human Ether-a-go-go Related Gene (HERG) channels expressed in xeopus oocytes and fleety cranking delayed therapy K currents of guinea gormandizer cardiomyocytes [17]. Kv4.3 underlies the molecular base of sub threshold cranking A-type Kv currents in the brain and the smooth muscles, and flash outside Kv currents in the heart [18]. Inhibition of CaMK II accelerated the inactivation of flash outside Kv current in mortal atrial myocytes and in murine colonic myocytes [19]. Indeed, the phosphorylation of Kv4.3 channels by CaMKII accelerated both the inactivation and the recovery from inactivation of Kv43 currents expressed in HEK cells, and the contrary effect was observed with the use of CaMKII impediments [20].
In silico computational protein – ligand docking has become a significant tool for medicine discovery and development of drugs and is generally initiated with structure identification of given target protein moieties of medical interest. Molecular docking is also used to dissect protein relations with seeker small moieties (ligands) according to conformations and binding free powers, which are expressed as ligand–protein list forces in kilocalories per mole (Kcal/mole) [21]. In a structure-grounded medicine design, molecular docking is one of the most generally used styles because of its capability to prognostic small-patch ligands list confirmation to the applicable target binding point [22]. As a direct and rational approach for medicine discovery, computational docking analysis establish virtual models of ligands – protein relation at the extremely small position and can be used to inform posterior confirmation using traditional in vitro and in vivo assays, therefore minimizing the time and cost of the drug discovery process [23]. Herein, we report in silico binding affinities of the three antipsychotic drugs CLZ, OLZ and TFZ using a set of open-source molecular docking programs.
Materials and Methods
Software and programs PyMol (DeLano Scientific LLC, Palo Alto, California, USA) and Discovery Studio Biovia 2017 (Dassault Systèmes, San Diego, California, USA) were employed to visualize and adapt receptor and ligand structures. AutoDock Vina (The Scripps Research Institute, La Jolla, San Diego, USA) was the primary docking program used in this study. The Survivin.PDBQT file format was prepared, and the grid box size was determined using AutoDock Tools version 1.5.4 (ADT; Scripps Research Institute, La Jolla, San Diego, USA). LigPlot+v.2.2-ligand-protein interaction diagrams (EMBL-EBI).
Preparation of macromolecule structures
The crystal structure of the PK’s was downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) protein data bank (http://www.rcsb.org) and changed into encoded with the distinctive PDB code (Table 1). Protein structure inhibitors had been separated via way of means of freeing atomic coordinates of the .PDB format. All water molecules had been removed, and ADT software program changed into used to put together the desired documents for AutoDock Vina via way of means of assigning hydrogen polaraties, calculating Gasteiger charges to protein structures, from the .PDB format to .PDBQT format [22].
| Different Protein kinases | PDB Id’s |
|---|---|
| AGC | 4UTD |
| CaM1 | 3KFX |
| CK1 | 4GHT |
| CMGC | 2IN6 |
| STE | 3DV3 |
| TK | 5B7V |
| Tyrosil –like kinases | 3RVG |
| Typical kinases | 5I35 |
Table 1: 3D structures model of different PK’s retrieved from Protein data with their PDB Id.
Preparation of ligand structures
Structures of CLZ, OLZ and TFZ were downloaded in the Spatial Data File (.SDF) file format from the PubChem Compound Database (National Center for Biotechnology Information; https://pubchem. ncbi.nlm.nih.gov/) (Figure 1) [24]. Physicochemical properties of the ligands met the standards of Lipinski’s rule of five, in any other case referred to as Lipinski’s rule of drug-likeness. Molecular weights and hydrogen-bonding interactions among donors and acceptors are important structural determinants of protein objectives and ligandbinding sites [25].
In particular, compounds are much more likely permeable and active as ligands when they have >5 hydrogen-bond donors, >10 hydrogen-bond acceptors, a molecular mass of >500, and calculated log P (CLog P) values of >5 [25]. Chemical structures in the .SDF format were converted to the .PDB format using Pymol. ADT was then used to analyze ligand structures in terms of combinations with non-polar hydrogen’s, additions of Gasteiger changes, and rotatable bonds. Structures in the ligand.PDB format were then converted to the ligand. PDBQT format using ADT, enabling use with AutoDock4 (AD4) and AutoDock Vina [26] (Table 2).
| Physicochemical properties | Clozapine | Olanzapine | Trifluoperazine |
|---|---|---|---|
| Class | Atypical | Atypical | Typical |
| Pubchem Id | 1.35E+08 | 1.35E+08 | 5566 |
| Systematic name | 8-chloro-11-(4-methylpiperazin-1yl)-5H-dibenzo[b,e] [1,4]diazepine | 2methyl- 4(4methyl-1-piperazinyl)10H thieno[2,3-b] [1,5]benzodiazepine | 10-[3-(4-methylpiperazine-1-yl)propyl]-2(trifluoromethyl)-10H- phenothiazine |
| Molecular formula | C17H20N4S | C17H20N4S | C21H24F3N3S |
| Molecular weight | 326.823 g/mol | 312.44 g/mol | 407.497 g/mol |
| Solubility | Ethanol | Methanol | Water |
| Hydrogen-binding donors | 1 | 1 | 0 |
| Hydrogen-binding acceptors | 4 | 4 | 4.5 |
| Log P | 3.235 | 3.562 | 4.923 |
Table 2: Physico-chemical properties of antipsychotic drugs retrieved from PubChem.
Docking Methodology
Molecular docking was performed using the Auto Dock Vina program. Ligands were docked individually to the receptor with grid coordinates (grid center) and grid boxes of certain sizes for each receptor [27]. The ligand was in a flexible condition when interacting with macromolecules under rigid conditions. The configuration file was engaged by opening notepad to run AutoDock Vina. ADT was required to prepare the input .PDBQT file for survivin and to set the size and the center of the grid box. Kollman charges and polar hydrogen atoms were included in the survivin structure. The grid size was set at 14 × 14 × 14 (x, y, and z) points, and the grid center was designated at x, y, and z dimensions respectively, with a grid spacing of 1000 A°. The prepared file was saved in the .PDBQT format. Ligand-binding affinities were predicted as negative Gibbs free energy (ΔG) scores (Kcal/mol), which were calculated on the basis of the AutoDock Vina scoring function (26).
Post-docking analyses were visualized using PyMOL and Discovery Studio Biovia 2017, and LigPlot which showed the sizes and locations of binding sites, hydrogen-bond interactions, hydrophobic interactions, and bonding distances as interaction radii of <5 A° from the position of the docked ligand. Subsequently, binding poses of each ligand were observed and their interactions with the protein were characterized, and the best and most energetically favorable conformations of each ligand were selected [28].
Results
PK’s have been concerned in wider range for cellular functions. Thus, these kinases are attractive targets for a variety of therapeutic approaches. Therefore, initially we have selected three FDA approved antipsychotics drugs CLZ, OLZ and TFZ which were used in in vitro experiments. In order to recognize an accurate docking routine for carrying out molecular docking studies of PK’s as well as to identify the potent inhibitors against that protein, Auto Dock Vina, Pymol, Discovery studio along with PLP (Protein Ligand Filters), Protein Plus and LigPlot was implicated.
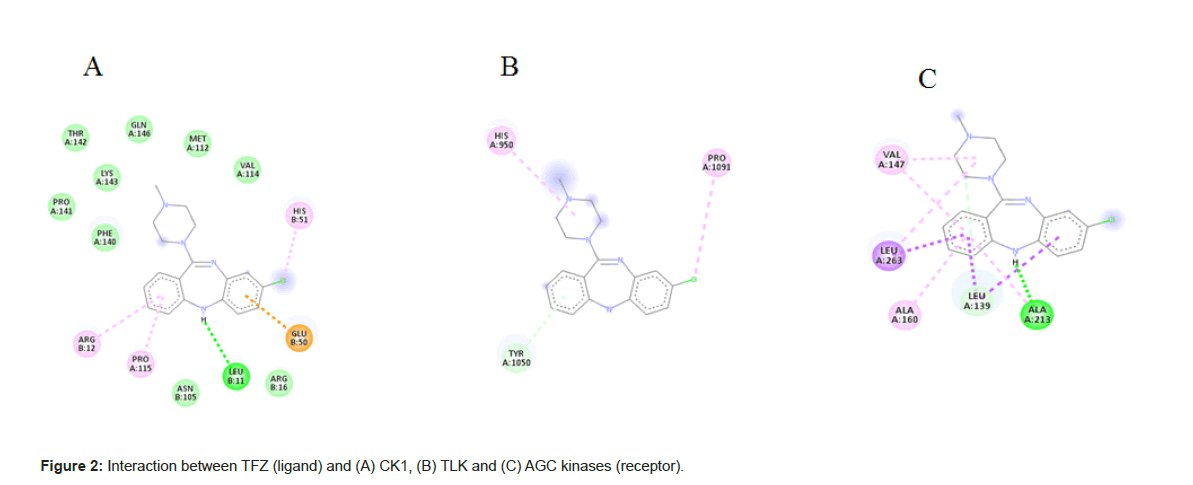
The 2D view of protein-ligandB interactions of the best poses generated by all the three antipsychotic drugs (Figure 2). The docking score of the antipsychotic drug varies from -9.1 Kcal mol-1 to -5.2 Kcal mol-1 (Table 3). In simultaneous, analyses, we mapped PK’s amino acid residues that are involved in hydrogen bond, electrostatic interactions with ligands using Auto Dock Vina.
| Different PK’s | ΔG = -Kcal mol−1 | ||
|---|---|---|---|
| Clozapine (CLZ) | Olanzapine (OLZ) | Trifluoperazine (TFZ) | |
| AGC | 8.5 | 7.5 | 8.8 |
| CaM1 | 6.4 | 6.5 | 6.8 |
| CK1 | 6.9 | 6 | 9.1 |
| CMGC | 8.4 | 5.5 | 8.1 |
| STE | 5.9 | 6.9 | 6 |
| TK | 8 | 6 | 8.1 |
| TLK | 6.5 | 5.3 | 9 |
| Typical Kinases | 5.5 | 5.2 | 5.9 |
Table 3: Binding free energy (ΔG) of different inhibitors with PK’s in – Kcal mol-1.
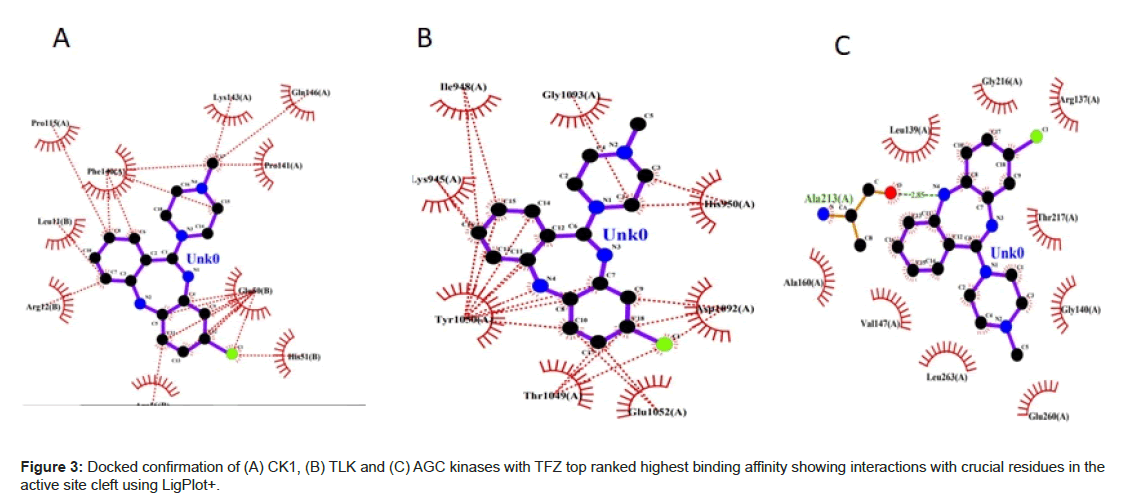
From the Figure 2A shows the binding mode of top pose CK1 in complexed with TFZ generated by using Discovery studio and LigPlot with the lowest docked binding affinities of -9.1 Kcal mol-1. TFZ formed three π-alkyl interactions from carbon of Arg 12 of chain B to benzene ring of TFZ at 5.47 A°, from pyrole of Pro 115 of chain B to benzene ring of TFZ at 5.43 A°, from imidazole ring of His 51 of chain B to fluorine of TFZ at 4.90 A°; TFZ forms one conventional hydrogen bond from oxygen of Leu 11 of chain B to hydrogen of thiazepene of TFZ 2.62 A° and one π-anion from oxygen of Glu 50 of chain B to benzene ring of TFZ at 3.71 A° (Figure 3A).
From the Figure 2B shows the binding mode of top pose TLK in complexed with TFZ with the lowest docked binding affinities of -9.0 Kcal mol-1. TFZ forms one alkyl bond from pyrole ring of Pro 1091 of chain A to fluorine of TFZ at 5.27 A°; forms one π-donor hydrogen bond from nitrogen of His 950 of chain A to diazene ring of TFZ at 3.82 A°; forms one π-π stacked interaction from phenol of Tyr 1050 of chain A to benzene ring of TFZ at 4.13 A° (Figure 3B).
From the Figure 2C shows the binding mode of top pose AGC kinases in complexed with the lowest docked binding affinities of -8.8 Kcal mol-1. TFZ forms one conventional hydrogen bond from oxygen of Ala 213 of chain A to diazepene ring of TFZ at 1.83 A°; forms one alkyl bond from carbon of Leu 263 of chain A to diazene ring of TFZ at 5.48 A°; forms one carbon hydrogenated bond from oxygen Leu 139 to piperazine ring of TFZ at 3.43 A°; forms four π-alkyl bond from carbon of Ala 213, Ala 160, Val 147 to benzene ring of TFZ at 5.06 A°, 4.54 A°, 5.13 A° and one from carbon of Val 147 to diazene ring of TFZ at 5.37 A° and three π-sigma bonds from carbon of Leu 139 of chain A to benzene ring of TFZ at 3.64 A° and 3.87 A° and one from Leu 263 of chain A to benzene ring of TFZ at 3.78 A° (Figure 3C). Docking of all the kinases with CLZ, OLZ and TFZ are enclosed in supplementary material.
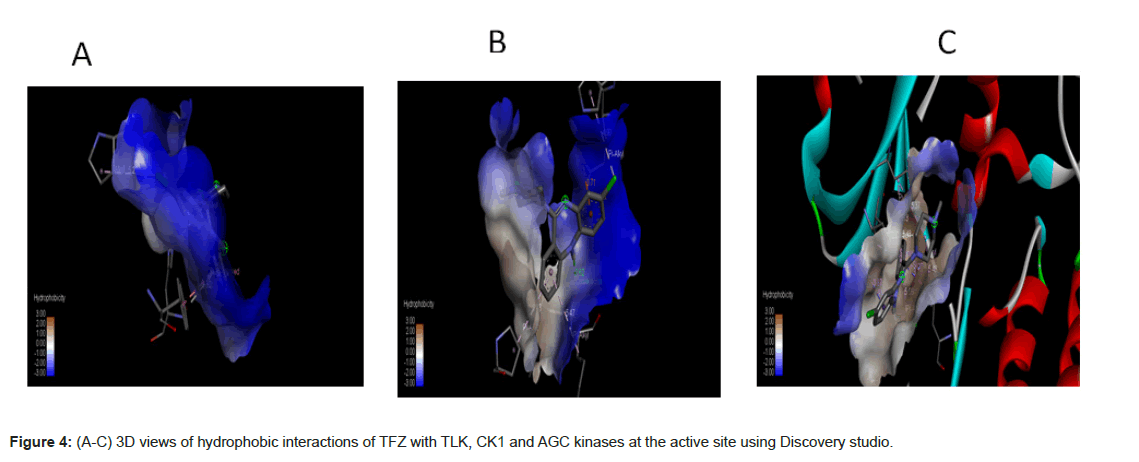
In the light of the above analysis, the CK1 docked posed generated by Auto Dock Vina produced the best results. It forms hydrogen bonds, hydrophobic interactions (Figure 4) with the important residues of the binding pocket of CK1 thus stabilizing the structure of target receptor. The dock pose with least binding energy has the highest affinity and hence is the best docked confirmation which could serve as lead molecule against the targeted protein.
Discussion
Molecular docking is a widely used, relatively fast, and economical computational tool for predicting in silico the binding modes and affinities of molecular recognition events and the concept of free energy (ΔG) is used to determine the binding affinity of protein-ligand complex in docking studies. The negative or low value of ΔG indicates the strong binding affinity between protein-ligand complexes and from our earlier investigations with molecular docking studies of antipsychotic drugs with protein phosphatases [29]. Hence, in the current study, the binding affinity was carried out and the determined binding free energy as shown in Table 3 which reflects the binding affinity of the different ligands to a PK’s by calculating intermolecular and torsional free energies using Auto Dock Vina.
More recently the antipsychotic drug TFZ has been used extensively to probe for CaM-dependent reactions. It is a phenothaizine derivative antipsychotic drug being observed as a PP-2B inhibitor [30]. TFZ has shown the highest binding affinity towards CK1 (-9.1 Kcal/mol) when compared to TLK (-9.0 Kcal/mol) and AGC (-8.8 Kcal/mol) followed by CMGC and TK (-8.1 Kcal/mol), CaM 1 (-6.8 Kcal/mol), STE (-6.0 Kcal/mol) and Typical kinases (-5.9 Kcal/mol). Free energies of TFZ of PK’s ranged from -9.1 to -5.9 kcal/mol.
However there is less/no study being carried out investigate the other popular antipsychotic drugs such as CLZ and OLZ with PK’s and also TFZ with other PK’s. In the present study, phenothiazine derivatives such as CLZ, OLZ, and TFZ were docked against different PK’s. Results indicate the relative binding affinity of phenothiazine derivatives i.e OLZ with different PP’s and mainly concentrating on AGC, STE, and CaM 1 kinases.
CLZ, a second generation antipsychotic drug strongly inhibits PP-2B activity, but had minimal effect on PP-1 and PP-2A activities at the concentrations studies. Therefore, docking studies of CLZ with different PK’s show that CLZ has higher binding affinity with AGC (-8.5 Kcal/mol) when compared to CMGC (8.4 Kcal/mol) and TK (8.0 Kcal/mol) followed by CK 1 (-6.9 Kcal/mol), TLK (-6.5 Kcal/mol) and CaM 1 (-6.4 Kcal/mol) and has least binding affinity with Typical kinases (-5.5 Kcal/mol).
OLZ has the highest binding affinity towards AGC (-7.5Kcal/ mol), when compared to STE (6.9 Kcal/mol) and CaM 1 (-6.5Kcal/ mol), followed by CK 1 and TK (-6.0 Kcal/mol) and CMGC (5.5Kcal/mol), TLK (-5.3 Kcal/mol) and least affinity towards Typical kinases (-5.2 Kcal/mol). The docking score for different PK’s nearly range from -9.1 Kcal/mol to -5.2 Kcal/mol which clearly indicates that the first generation drug TFZ has more binding affinity compared to other drugs with PK’s.
The present study is done to predict the binding affinity of antipsychotic drugs with different PK’s in a view to discover potential therapeutic strategy in silico approach which can provide significant insights into other major characteristics of antipsychotic drugs and their binding with the PK’s. Henceforth, the results obtained would lead us to know the different roles of antipsychotic drugs and their interactions with different PK’s. This can be further used as potential targets in the field of drug designing.
Conclusion
Summarized record at the kinase antagonist drug design shows a massive step forward from the preliminary structure based design (performed on the basis of the first X-ray analysis in 1991) to emergence of modern concepts in this field. The modern drug design cannot be imagined without docking procedure even if knowledge about a 3D spatial structure is limited. The 3D structure assemblage and a profound insight into kinase interaction patterns have advanced kinase drug development to the next step of understanding. Nevertheless, successful design of the in silico inhibitor as potential drug depends on a variety of conditions such as: the correct choice of the validated kinase target, availability of the 3D structure information about specific kinase-inhibitor interactions, at least one high X-ray resolution algorithm.
In this study, these inhibitors were re-docked in protein complex using docking score, hydrogen bond and hydrophobic interactions, and binding free energy. All the docking complexes have better docking scores and exhibited good binding free energy. Hydrogen and hydrophobic interactions were analyzed and revealed that having exhibited favorable interactions with the active site residues. Among all the docking scores TFZ showed the highest binding affinity towards CK1 (-9.1 Kcal mol-1), followed by TLK (-9.0 Kcal mol-1) and AGC kinases (-8.8 Kcal mol-1) when compared to CLZ and OLZ. The potential inhibition of TFZ towards PK’s could be revealed through in vivo and in vitro studies. Therefore, TFZ drug could serve as chief molecule against the targeted protein.
References
- Urich R, Wishart G, Kiczun M, Richters A, Tidten-Luksch N et al. (2013) De Novo Design of Protein Kinase Inhibitors by in Silico Identification of Hinge Region-Binding Fragments. ACS Chem Biol 8(5): 1044–1052.
[Crossref] [Google scholar][Pubmed]
- Dubinina GG, Chupryna OO, Platonov MO, Borisko PO, Ostrovska GV et al. (2007) In Silico Design of Protein Kinase Inhibitors: Successes and Failures. Anticancer Agents Med Chem 7(2):171–188.
[Crossref] [Google scholar][Pubmed]
- Minuesa G, Albanese SK, Xie W, Kazansky Y, Worroll D et al.(2019)Small-Molecule Targeting of MUSASHI RNA-Binding Activity in Acute Myeloid Leukemia. Nat Commun 10 (1):2691.
[Crossref] [Google scholar][Pubmed]
- Pushparaj JR , Moorthiraman M , Sarangapani B , Rajaram R (2021) Molecular Docking Performance of Selective Organic Compounds with Target Protein. Biointerface Res Appl Chem 11 (4): 12414–12424.
- Qi L, Ding Y (2013) Potential Antitumor Mechanisms of Phenothiazine Drugs . Sci China Life Sci 56 (11): 1020–1027.
[Crossref] [Google scholar][Pubmed]
- Rimessi A, Pavan C, Ioannidi E, Nigro F, Morganti C et al. (2017) Protein Kinase C β: A New Target Therapy to Prevent the Long-Term Atypical Antipsychotic-Induced Weight Gain. Neuropsychopharmacol 42 (7):1491–1501.
[Crossref] [Google scholar][Pubmed]
- Yadav PN, Abbas AI, Farrell MS, Setola V, Sciaky N et al. (2011) The Presynaptic Component of the Serotonergic System Is Required for Clozapine’s Efficacy. Neuropsychopharmacol 36 (3):638–651.
[Crossref] [Google scholar][Pubmed]
- Birjandi AA, Suzano FR, Sharpe P T (2020) Drug Repurposing in Dentistry: Towards Application of Small Molecules in Dentin Repair. Int J Mol Sci 21 (17): 6394.
[Crossref] [Google scholar][Pubmed]
- Mondelli V, Anacker C, Vernon AC, Cattaneo A, Natesan S et al. (2013) Haloperidol and Olanzapine Mediate Metabolic Abnormalities through Different Molecular Pathways. Transl Psychiatry 3(1):e208–e208.
[Crossref] [Google scholar][Pubmed]
- Howland RH (2016) Trifluoperazine: A Sprightly Old Drug. J Psychosoc Nurs Ment Health Serv 54 (1): 20–22.
[Crossref] [Google scholar][Pubmed]
- Xia Y, Jia C, Xue Q, Jiang J, Xie Y et al. (2019) Antipsychotic Drug Trifluoperazine Suppresses Colorectal Cancer by Inducing G0/G1 Arrest and Apoptosis. Front Pharmacol 10: 1029.
[Crossref] [Google scholar][Pubmed]
- Feng Z, Xia Y, Gao T, Xu F, Lei Q et al. (2018)The Antipsychotic Agent Trifluoperazine Hydrochloride Suppresses Triple-Negative Breast Cancer Tumor Growth and Brain Metastasis by Inducing G0/G1 Arrest and Apoptosis. Cell Death Dis 9 (10): 1006.
[Crossref] [Google scholar][Pubmed]
- Li A, Chen X, Jing Z, Chen J (2020) Trifluoperazine Induces Cellular Apoptosis by Inhibiting Autophagy and Targeting NUPR1 in Multiple Myeloma. FEBS Open Bio 10 (10): 2097–2106.
[Crossref] [Google scholar][Pubmed]
- Wang B, Luo Y, Zhou X, Li R (2018) Trifluoperazine Induces Apoptosis through the Upregulation of Bax/Bcl‑2 and Downregulated Phosphorylation of AKT in Mesangial Cells and Improves Renal Function in Lupus Nephritis Mice. Int J Mol Med 41(6): 3278-3286.
[Crossref] [Google scholar][Pubmed]
- Zhelev Z, Ohba H, Bakalova R, Hadjimitova, Ishikawa M et al. (2004) Phenothiazines Suppress Proliferation and Induce Apoptosis in Cultured Leukemic Cells without Any Influence on the Viability of Normal Lymphocytes. Cancer Chemother. Pharmacol 53 (3):267-275.
[Crossref] [Google scholar][Pubmed]
- Hong DH, Son YK, Li H, Jung ID, Park YM et al. (2014) The Calmodulin Inhibitor and Antipsychotic Drug Trifluoperazine Inhibits Voltage-Dependent K+ Channels in Rabbit Coronary Arterial Smooth Muscle Cells. Biochem Biophy Res Commun 443 (1): 321-325.
[Crossref] [Google scholar][Pubmed]
- Choi SY, Koh YS, Jo SH (2005) Inhibition of Human Ether-a-Go-Go -Related Gene K + Channel and I Kr of Guinea Pig Cardiomyocytes by Antipsychotic Drug Trifluoperazine. J Pharmacol Exp Ther 313 (2): 888-895.
- Dixon JE, Shi W, Wang HS, McDonald C, Yu H et al. (1996) Role of the Kv4.3 K + Channel in Ventricular Muscle: A Molecular Correlate for the Transient Outward Current. Circ Res 79 (4): 659-668.
[Crossref] [Google scholar][Pubmed]
- Tessier S, Karczewski P, Krause EG, Pansard Y, Acar C et al.(1999) Regulation of the Transient Outward K + Current by Ca 2+ /Calmodulin-Dependent Protein Kinases II in Human Atrial Myocytes. Circ Res 85 (9): 810–819.
[Crossref] [Google scholar][Pubmed]
- Chae YJ, Choi BH, Choi JS , Hahn SJ(2014) Block of Kv4.3 Potassium Channel by Trifluoperazine Independent of CaMKII. Neurosci Lett 578: 159–164.
[Crossref] [Google scholar][Pubmed]
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS et al. (2016)Computational Protein–Ligand Docking and Virtual Drug Screening with the AutoDock Suite. Nat Protoc 11 (5): 905–919.
[Crossref] [Google scholar][Pubmed]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK et al. (2009) AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem 30 (16): 2785–2791.
[Crossref] [Google scholar][Pubmed]
- Meng XY, Zhang HX, Mezei M, Cui M (2011)Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr Comput Aided-Drug Des 7 (2): 146–157.
[Crossref] [Google scholar][Pubmed]
- Sunagar MG, Aravind P, Gaonkar S, Devaraju KS, Joshi SD et al. (2018) In Silico Binding Affinity Studies of N-9 Substituted 6-(4-(4-Propoxyphenyl)Piperazin-1-Yl)-9H-Purine Derivatives-Target for P70-S6K1 & PI3K-δ Kinases. J Basic Appl Sci 7 (1): 84–91.
- Lipinski CA (2016) Rule of Five in 2015 and beyond: Target and Ligand Structural Limitations, Ligand Chemistry Structure and Drug Discovery Project Decisions. Adv Drug Deliv Rev 101:34–41.
[Crossref] [Google scholar][Pubmed]
- Trott O, Olson AJ (2009) AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem 31(2): 455–461.
[Crossref] [Google scholar][Pubmed]
- Harish BM, Saraswathi R, Vinod D, Devaraju KS (2016) Discovery of a Latent Calcineurin Inhibitory Peptide from Its Autoinhibitory Domain by Docking, Dynamic Simulation, and in Vitro Methods. J Biomol Struct Dyn 34 (5): 983–992.
[Crossref] [Google scholar][Pubmed]
- Afriza D, Suriyah WH, Ichwan SJA (2018) In Silico Analysis of Molecular Interactions between the Anti-Apoptotic Protein Survivin and Dentatin, Nordentatin, and Quercetin. J Phys Conf Ser 1073 (3): 1-7
- Devaraju KS, Bulbule SR , Aravind P (2023) In silico Binding affinity studies of Antipsychotic drugs with Protein Phosphatases by Molecular Docking . J Appl Bioinforma Comput Biol 31(1):1
- Feldkamp MD, Gakhar L, Pandey N, Shea MA (2015) Opposing Orientations of the Anti-Psychotic Drug Trifluoperazine Selected by Alternate Conformations of M144 in Calmodulin: To Bury or Expose TFP CF 3 : CaM Met144 Decides. Proteins Struct Funct Bioinforma 83 (5): 989-996.
[Crossref] [Google scholar][Pubmed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi