Research Article, Med Microbiol Rep Vol: 7 Issue: 1
Correlation of Gram Staining and Quantitative Cultures of Endo- Tracheal Aspirates in Mechanically Ventilated Patients: An Experience from Tertiary Care Hospital
Heena Tak, Kanne Padmaja* and Sukanya Sudhaharan
Department of Microbiology, Nizams Institute of Medical Sciences, Telangana, India
*Corresponding Author: Kanne Padmaja
Department of Microbiology, Nizams Institute of Medical Sciences, Telangana, India
Tel: 8639918215
E-mail: kannepadmaja@gmail.com
Received date: 04 December 2022, Manuscript No. MMR-22-82230;
Editor assigned date: 07 December 2022, PreQC No. MMR-22-82230 (PQ);
Reviewed date: 21 December 2022, QC No. MMR-22-82230;
Revised date: 06 February 2023, Manuscript No. MMR-22-82230 (R);
Published date: 13 February 2023, DOI: 10.4172/ MMR.1000326
Citation: Tak H, Padmaja K, Sudhaharan S (2023) Correlation of Gram Staining and Quantitative Cultures of Endo-Tracheal Aspirates in Mechanically Ventilated Patients: An Experience from Tertiary Care Hospital. Med Microbiol Rep 7:1.
Abstract
Introduction: Ventilator Associated Pneumonia (VAP) is the most common and fatal nosocomial infection in Intensive Care Units (ICU). VAP is associated with prolonged hospital and ICU stay, high hospital costs, and increased mortality. The accurate diagnosis of VAP causes great difficulty and remains a constant challenge for clinical practice.
Purpose: To determine the effectiveness of gram staining of Endo-Tracheal Aspirates (ETA) and quantitative cultures for predicting VAP and to study the microbiology and susceptibility patterns of MDR isolates.
Methodology: Data was retrieved retrospectively through the records from Jan 1st 2016-July 31st). A total of 865 ETA received from different ICUs were included in the study. Gram staining and quantitative culture results were analyzed. The diagnostic thresholds for ETA quantitative cultures were taken as 105 cfu/ml. Growth below the threshold was assumed to be due to colonization. Diagnostic threshold for gram stain results was assumed as >103.
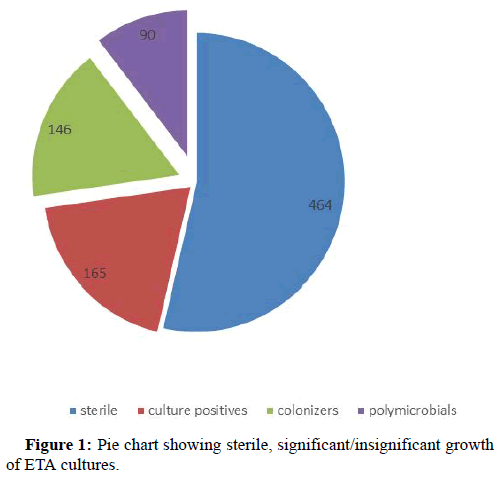
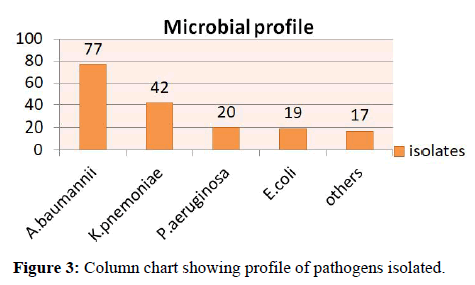
Results: Out of total 865 samples 464 were sterile, 255 were culture positive and 146 were colonizers and 90 were polymicrobial. Acinetobacter baumannii (44.31%) was found to be the commonest pathogen isolated followed by Klebsiella pneumoniae (23.86), Pseudomonas spp. (11%), Escherichia coli (10.22%). 84.09% of isolates were multi drug resistant. The sensitivity of our study was 75%, specificity 53.42% with a positive predictive value of 70.9%and negative predictive value of 58.90%.
Conclusion: Our study emphasizes that an inevitable oropharyngeal bacterial contamination occurs during the collection of endotracheal aspirates, quantitative culture techniques are always needed to differentiate oropharyngeal contaminants present at low concentration from the infective organisms that are likely to be the cause of pneumonia.
Keywords: Gram staining; Ventilator-associated pneumonia; Mechanical ventilation; Endo-tracheal aspirate; Bacterial culture
Introduction
Critically ill patients in Intensive Care Units (ICU) are at higher risk of contracting nosocomial infections, hence complicating the primary disease process resulting in high morbidity, mortality, treatment failure, and increased hospital stay and cost [1]. Pneumonia is the second most common nosocomial infection in critical patients, affecting 27% of all critically ill patients [2]. Majority of nosocomial pneumonias are attributed to presence of mechanical ventilation and is called as Ventilator-Associated Pneumonia (VAP) [3]. VAP is defined as pneumonia that occurs 48 h or more after endotracheal intubation or tracheostomy, caused by microorganism that was not present or incubating at the time mechanical ventilation was started. It can be of two types;
• Early-onset VAP which is defined as VAP that occurs within the
first 4 days of ventilation.
• Late-onset VAP which is defined as VAP that occurs after 4 days of
ventilation.
Depending on the surveillance methods used for the diagnosis of VAP, the risk of this complication ranges from 1.2 to 8.5 cases per 1,000 ventilator-days (0.6%-4%) [4]. The mortality attributable to VAP has been reported to range between 0% and 50% [5]. VAP is preventable complication, timely and appropriate diagnosis is important in guiding antimicrobial therapy and decreasing mortality, morbidity and health care cost associated with it. Various severity scoring criteria like Clinical Pulmonary Scoring System (CPIS), modified CPIS, lung ultrasound and pentraxin-3 pulmonary score, etc., have been formulated for accurately diagnosing VAP in clinical settings, they include clinical picture, imaging techniques, microbiological analysis for samples obtained, and biomarkers of host response. Microbiological diagnosis of VAP includes gram staining and subsequent culture methods of respiratory tract secretions obtained using either endotracheal aspiration or fiberoptic bronchoscopy [6]. Culture methods of respiratory tract secretions obtained by various defined methods are considered as gold standard for confirming the causative microorganism; however the Turnaround Time (TAT) for identification and antimicrobial susceptibility takes at least 48 hours-72 hours. Gram stain of respiratory specimens can provide quick preliminary report of suspected bacterial pathogen. The effectiveness of the gram stain of endotracheal aspirate for diagnosing VAP was found useful by several studies [7,8]. In this study, we assessed effectiveness and correlation of gram staining and quantitative culture in diagnosing VAP.
Aims and objectives
• To determine the effectiveness of gram staining of endo-tracheal
aspirates and quantitative cultures for predicting VAP.
• To study the microbiology and susceptibility patterns of multi drug
resistant isolates.
Materials and Methods
A retrospective study was undertaken from 1st January 2016-31st July 2019 at Nizams institute of medical sciences, Hyderabad. All the required clinical data was retrieved retrospectively through the records. A total of 865 endo-tracheal aspirates received from different ICUs were included in the study. The clinical samples included in the study was endo tracheal aspirates conventionally processed by standard laboratory protocol (as per CMPH guideline) at microbiology laboratory.
Microbiology workup
Endo-tracheal aspirate specimens were quantitatively (neat dilution, 1:10, 1:100 and 1:1000 dilution) cultured on chromogenic agar and 5% sheep blood agar (biomeriux, France, Marcy’l Etoile) and incubated in aerobic conditions at 37°C for 18 hours-24 hours. The gram smear was prepared from the neat dilution. Endotracheal aspirate cultures showing growth of ≥ 105 CFU/ml in microbiology are considered as significant growth (laboratory proven VAP) while cultures with growth ≤ 105 CFU/ml are considered as colonizers. Identification and susceptibility testing of isolates from significant growth was done by using Vitek 2 compact system. IDGN, N280 and N281 panels were used for identification of gram-negative pathogens and IDGP and P628 panels were used for identification of gram positive pathogens. The microscopic threshold for diagnosis of VAP with endo tracheal aspirates >10 Polymorphonuclear Neutrophils (PMN)/High-Power Field (HPF), ≥ 1 bacteria/Oil Immersion Field (OIF).
Results
A total of 865 ETA samples were received from different ICUs across the hospital by the department of microbiology from January 1st 2016-July to 31st 2019. Out of 865 samples 464 ETA samples were sterile after overnight incubation. On further evaluation of remaining 401 (865-464) ETA samples, 90 samples were showing more than 3 different organisms indicating possible contamination during sample collection, while remaining 311 samples included 165 samples had significant growth of ≥ 105 CFU/ml and 146 samples had insignificant growth (<105 CFU/ml), hence were considered as colonizers (Figure 1).
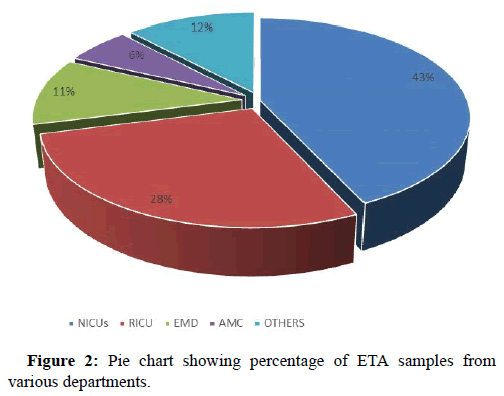
On analyzing the data it was observed that major chunk of ETA samples were received from neurology ICUs 375 out of 865 that included 237 samples from Neurosurgery ICU (NSICU) and 138 from neuro-medical (NLICU), followed by Respiratory Intermediate Care Unit (RICU) n=242, Emergency Department (EMD) n=95, Acute Medical Care (AMC) n= 51 and rest of the samples were from other departments of our institute (Figure 2).
Of 165 ETA samples with significant growth of ≥ 105 CFU/ml, 10 samples were having significant growth of 2 microorganisms while remaining were having monomicrobial growth.
Gram smear of 311 samples were analyzed that included 165 ETA samples with significant growth and 146 ETA samples with insignificant growth. For a total of 192 ETA samples gram smear were showing plenty of polymorphs with and without micro-organisms taking gram staining thresholds for diagnosis of VAP with ETA: >10 Polymorphnuclear Neutrophils (PMN)/High-Power Field (HPF), ≥ 1 bacteria/oil immersion field (OIF). Positive gram smear findings and culture positivity (significant growth) observed in 124 ETA samples whereas gram smear were negative for 41 samples with significant growth. However, for 68 of the ETA samples with insignificant growth, gram smear findings were positive (Table 1).
| Gramsmear | Correlation | ||
|---|---|---|---|
| True positive | Gram +ve | Significant growth | 124 |
| True negative | Gram -ve | insignificant growth | 78 |
| False positive | Gram +ve | insignificant growth | 68 |
| False negative | Gram –ve | Significant growth | 41 |
Table 1: Showing gram smear and culture finding/correlation.
The sensitivity of gram smear was observed in our study was 75% whereas specificity as low as 53.42%.
CPIS scoring of 311 ETA samples (significant growth samples and insignificant growth samples) were analyzed for clinical presence and absence of VAP by retrospectively reviewing the patient case files. Of 165 samples with significant growth 160 corresponding patients were diagnosed with clinical VAP whereas for remaining 5 samples with significant growth did not have clinical VAP. 130 samples with insignificant growth were in line with clinical absence of VAP, however 16 ETA samples with insignificant growth were having clinical VAP (Table 2).
| Correlation | Culture findings | Clinical VAP | |
|---|---|---|---|
| True positive | Culture ≥ 1 lakh cfu/ml | VAP present | 160 |
| True negative | Culture < 1 lakh cfu/ml | VAP absent | 130 |
| False positive | Culture ≥ 1 lakh cfu/ml | VAP absent | 5 |
| False negative | Culture < 1 lakh cfu/ml | VAP present | 16 |
Table 2: Showing correlation between culture findings and clinical VAP.
The sensitivity and specificity of quantitative culture of ETA was observed as 90.91% and 97.7% respectively for lab. diagnosis of VAP. The positive predictive value of quantitative culture of ETA was found to be 96.97% and negative predictive value was observed as 89.04%.
Microbial profile of pathogenic isolates from samples with significant growth were analyzed, Acinetobacter baumannii was found to be the most common isolate 44.31% (n=77), followed by Klebsiella pneumonia 23.86% (n=42), Pseudomonas aeruginosa, 11% (n=20), Escherichia coli 10.22% (n=19) I and others like, Enterococcus spp. 4% (n=7), Staphylococcus aureus 3.8% (n=6) and Serratia marcescens 2.2% (n=4) (Figure 3).
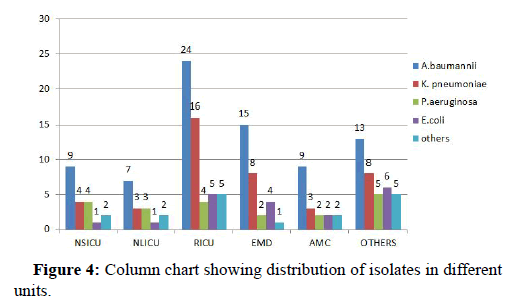
30.85% (n=54) pathogenic isolates were from RICU followed by EMD 17.14% (n=30), NSICU accounted for 11.42% (n=20), AMC 10.28% (n=18), NLICU 9.14% (n=16) and remaining 37.21% (n=37) from other departments (Figure 4).
Antimicrobial sensitivity of the pathogenic isolates were analysed for both groups of isolates (gram positive and gram negative). Highest drug resistance was observed in Acinetobacter baumannii, followed by. K. pneumonia. All A. baumannii isolates were found resistant to cephalosporins whereas only 1.5% of them were resistant to tigecyclin and colistin where as 4.7% of K. pneumonia isolates were also resistant to tigecyclin and colistin. The resistance pattern of remaining isolates in shown in Tables 3 and 4.
| Antibiotics | Acinetobacter baumannii (n=77) | Klebsiella pneumonia (n=42) | Pseudomonas aeruginosa (n=20) | Escherichia coli (n=19) | Serratia spp. (n=4) |
|---|---|---|---|---|---|
| Cefepime | 100% | 85.70% | 28.50% | 68.40% | 75% |
| Ceftazidime | 100% | 85.70% | 28.50% | 73.60% | 75% |
| Amikacin | 74.04% | 80.90% | 90% | 68.40% | 75% |
| Gentamicin | 93.50% | 80.90% | 57.14% | 68.40% | 75% |
| Meropenem | 90.90% | 78% | 65% | 52.60% | 75% |
| Doripenem | 90.90% | 78% | 65% | 52.60% | 75% |
| Levofloxacin | 94.80% | 85.70% | 90% | 84.20% | 75% |
| Cotrimoxazole | 94.80% | 70% | Intrinsic resistance | 68.40% | 75% |
| Colistin | 1.20% | 4.70% | 0 | 0 | Intrinsic resistance |
| Tigecycline | 1.20% | 4.70% | Intrinsic resistance | 0% | 75% |
Table 3: Showing resistance pattern of gram negative isolates.
| Antibiotic | Enterococcus Spp. n=7 | Staphylococcus aureus n=6 |
|---|---|---|
| Benzlypenicillin | 18.18% | 83.3%% |
| Oxacillin | not tested | 66.60% |
| Gentamicin | not tested | 66.60% |
| Ciprofloxacin | 9.00% | 33.33% |
| Levofloxacin | 18.80% | 33.33% |
| Clindamycin | not tested | 16.66% |
| Eythromycin | 18.80% | 33.33% |
| Linezolid | 100% | 100% |
| Daptomycin | not tested | 100% |
| Teicoplanin | 100% | 100% |
| Vancomycin | 100% | 100% |
| Tetracycline | 100% | 66.66% |
| Tigecycline | 100% | 100% |
| TMP/SMX | not tested | 66.66% |
Table 4: Resistance pattern of gram positive patterns.
Discussion
VAP is a preventable fatal nosocomial infection that occur in subset ICU patients, on mechanical ventilation. Diagnostic challenges in clinical settings remains a constant matter of concern. In this study we analyzed the gram staining and quantitative cultures of ETA in predicting VAP along with other components of CPIS scoring system. Gram staining, a potentially useful aid to help to guide empirical antimicrobial therapy in patients with VAP as it can provide immediate information about causative organisms. However, in our study the sensitivity of gram staining was found 75% whereas specificity was as low as 53.43%. A study from Zurich has reported sensitivity as high as 96% but specificity was found 12% [9]. In another study from France sensitivity and specificity was found 95% and 61% respectively [10]. In our study the sensitivity of quantitative ETA culture was found 90.91% and specificity 97.7%. In a study by Luis Fernando et al., reported sensitivity and specificity of 68% and 48% respectively [11]. In a study from India by Noyal M Joseph, et al., the sensitivity and specificity was found 88.1% and 84.2% respectively [12]. Due to their high specificity rate, quantitative cultures continue to be the gold standard. In this study we found that gram negative organisms are more prevalent accounting for 92.5% cases of VAP while gram positive pathogens caused only 7.5% of cases. Our findings are similar to the study done by Zorana M Djordjevic et al., where gram negative organisms accounted for above 90% cases [13]. In a study by Su Young Chi et al., from South Korea, gram positive pathogens (S. aureus) was found more common than gram negative in VAP [14].
Acinetobacter species are common cause of VAP. Since this organism survives in moist and dry conditions for a prolonged period, it often leads to nosocomial outbreaks. In our study Acinetobacter baumannii accounted for 44.31% (n=77), followed by Klebsiella pneumonia 23.86% (n=42), Pseudomonas aeruginosa 11% (n=20), Escherichia coli 10.22% (n=19) I and others like, Enterococcus spp. 4% (n=7), Staphylococcus aureus 3.8% (n=6) and Serratia marcescens 2.2% (n=4). In a study from India by Yogesh Harde, et al., the pathogenic profile was almost like our study, with Acinatobacter baumannii being most common [15]. Pathogenic spectrum of a study by Zorana M. Djordjevic, et al. has minor variation in comparison to our study, in both studies Acinatobacter baumannii was found most common pathogen but in later study Pseudomonas spp. was second common pathogen while it was Klebseialla in our study. Baraibar, et al., has reported only 8.1% of VAP due to Acinatobacter baumannii in her study which are far less in number than our study [16]. Likewise in a study by Mandakini Pawar, et al., the Acinatobacter baumannii was reported in 16% of cases of VAP [17]. In later study the gram positive pathogens accounted for 16% and no VAP was reported due to fungi, these findings are quite in line with our findings. In our study 6 (3.4%) isolates were S. aureus and all of them were sensitive to methicillin whereas in a study by Alok Gupta, el al., 8 (23.52%) isolates were S. aureus however in later study all S. aureus isolates were resistant to methicillin while none of our isolate was resistant to methicillin [18].
The menace of rising antimicrobial resistance in hospital set up across the globe is making treatment failure more prevalent and is also acting as an additional risk factor for increased mortality among critically ill patients. VAP due to a Multi Drug Resistant (MDR) organism is one of the serious complications. Studies have suggested that risk of death is more in patients who have VAP due to a multidrug-resistant pathogen. In our study highest drug resistance was observed in Acinetobacter baumannii that agrees with study by Zorana M. Djordjevi, et al.
Conclusion
We conclude that gram staining and qualitative cultures have fair correlation, in predicting VAP. Quantitative cultures can aid in differentiating a true pathogen from colonizers. Timely and accurate diagnosis can help in choosing right antimicrobial therapy and can prevent the irrational use of antibiotics.
References
- Klompas M (2007) Does this patient have ventilator-associated pneumonia? JAMA 297:1583-1593.
[Crossref] [Google Scholar] [PubMed]
- Papazian L, Klompas M, Luyt CE (2020) Ventilator-associated pneumonia in adults: A narrative review. Intensive Care Med 46:888-906.
[Crossref] [Google Scholar] [PubMed]
- Koenig SM, Truwit JD (2006) Ventilator-associated pneumonia: Diagnosis, treatment, and prevention. Clin Microbiol Rev 19:637-657.
[Crossref] [Google Scholar] [PubMed]
- Skrupky LP, McConnell K, Dallas J, Kollef MH (2012) A comparison of ventilator-associated pneumonia rates as identified according to the national healthcare safety network and American college of chest physicians criteria. Crit Care Med 40:281-284.
[Crossref] [Google Scholar] [PubMed]
- Ranjan N, Chaudhary U, Chaudhry D, Ranjan KP (2014) Ventilator-associated pneumonia in a tertiary care intensive care unit: Analysis of incidence, risk factors and mortality. Indian J Crit Care Med 18:200-204.
[Crossref] [Google Scholar] [PubMed]
- Hashimoto S, Shime N (2013) Evaluation of semi-quantitative scoring of gram staining or semi-quantitative culture for the diagnosis of ventilator-associated pneumonia: A retrospective comparison with quantitative culture. J Intensive Care 1:1-5.
[Crossref] [Google Scholar] [PubMed]
- O'Horo JC, Thompson D, Safdar N (2012) Is the gram stain useful in the microbiologic diagnosis of VAP? A meta-analysis. Clin Infect Dis 55:551-561.
[Crossref] [Google Scholar] [PubMed]
- Brun-Buisson C, Fartoukh M, Lechapt E, Honore S, Zahar JR, et al. (2005) Contribution of blinded, protected quantitative specimens to the diagnostic and therapeutic management of ventilator-associated pneumonia. Chest 128:533-544.
[Crossref] [Google Scholar] [PubMed]
- Chuard C, Reller LB (1999) Diagnostic value of gram stain and culture of sputum and endotracheal aspirates in bacteremic pneumococcal pneumonia. Clin Microbiol Infect 5:106-109.
[Crossref] [Google Scholar] [PubMed]
- Blot F, Raynard B, Chachaty E, Tancrede CY, Antoun S, et al. (2000) Value of gram stain examination of lower respiratory tract secretions for early diagnosis of nosocomial pneumonia. Am J Respir Crit Care Med 162:1731-1737.
[Crossref] [Google Scholar] [PubMed]
- Camargo LF, de Marco FV, Barbas CS, Hoelz C, Bueno MA, et al. (2004) Ventilator associated pneumonia: Comparison between quantitative and qualitative cultures of tracheal aspirates. Crit Care 8:1-9.
[Crossref] [Google Scholar] [PubMed]
- Joseph NM, Sistla S, Dutta TK, Badhe AS, Rasitha D, et al. (2010) Role of semi-quantitative and quantitative cultures of endotracheal aspirates in the diagnosis of ventilator-associated pneumonia. Aust Med J 3:627.
- Djordjevic ZM, Folic MM, Jankovic SM (2017) Distribution and antibiotic susceptibility of pathogens isolated from adults with hospital-acquired and ventilator-associated pneumonia in intensive care unit. J Infect Public Health 10:740-744.
[Crossref] [Google Scholar] [PubMed]
- Chi SY, Kim TO, Park CW, Yu JY, Lee B, et al. (2012) Bacterial pathogens of ventilator associated pneumonia in a tertiary referral hospital. Tuberc Respir Dis (Seoul) 73:32-37.
[Crossref] [Google Scholar] [PubMed]
- Harde Y, Rao SM, Sahoo J, Bharuka A, Swetha B, et al. (1926) Detection of ventilator associated pneumonia, using Clinical Pulmonary Infection Score (CPIS) in critically ill neurological patients. Mortality. 84:1-96.
- Baraibar J, Correa H, Mariscal D, Gallego M, Valles J, et al. (1997) Risk factors for infection by Acinetobacter baumannii in intubated patients with nosocomial pneumonia. Chest 112:1050-1054.
[Crossref] [Google Scholar] [PubMed]
- Pawar M, Mehta Y, Khurana P, Chaudhary A, Kulkarni V, et al. (2003) Ventilator-associated pneumonia: Incidence, risk factors, outcome, and microbiology. J Cardiothorac Vasc Anesth. 17:22-28.
[Crossref] [Google Scholar] [PubMed]
- Gupta A, Agrawal A, Mehrotra S, Singh A, Malik S, et al. (2011) Incidence, risk stratification, antibiogram of pathogens isolated and clinical outcome of ventilator associated pneumonia. Indian J Crit Care Med 15:96-101.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi