Research Article, J Appl Bioinforma Comput Biol Vol: 10 Issue: 5
Allele Frequency Associated With Dhfr And Dhps Genes Obtained From Plasmodium vivax In Anambara State, Nigeria
Isaac Okezie Godwin1, Ifeoma Mercy Ekejindu2, George Uchenna Eleje3, Alfred Friday Ehiaghe4, Bright Chukwuebuka Unaeze5 and George Oche Ambrose6*
1Department of Medical Laboratory Science, Faculty of Health Sciences and Technology, Nnamdi Azikiwe University, Nnewi Campus, Nnewi, Anambra State, Nigeria
2Department of Medical Laboratory Science, Faculty of Health Sciences and Technology, Nnamdi Azikiwe University, Nnewi Campus, Nnewi, Anambra State, Nigeria
3Department of Obstetrics and Gynaecology, Nnamdi Azikiwe University Teaching Hospital, Nnewi, Anambra State, Nigeria
4Department of Medical Laboratory Science, Faculty of Health Sciences and Technology, Nnamdi Azikiwe University, Awka, Anambra State, Nigeria
5Department of Medical Laboratory Science, Faculty of Health Sciences and Technology, Nnamdi Azikiwe University, Awka, Anambra State, Nigeria
6Centre for Malaria and other Tropical Diseases, University of Ilorin Teaching Hospital, Kwara State, Nigeria
*Corresponding Author: George Oche Ambrose
Centre for Malaria and other Tropical Diseases, University of Ilorin Teaching Hospital, Kwara State, Nigeria
E-mail: Ocheab1@gmail.com
Received: April 16, 2021 Accepted: May 11, 2021 Published: May 18, 2021
Citation: Godwin IO, Ekejindu IM, Eleje GU, Ehiaghe AF, Unaeze BC, et al. (2021) Allele Frequency Associated With Dhfr And Dhps Genes Obtained From Plasmodium Vivax In Anambara State, Nigeria. J Appl Bioinforma Comput Biol 10:5.
Abstract
The Plasmodium vivax dihydrofolate reductase (DHFR) and dihydropteroate synthetase (DHPS) are enzymes of central importance in parasite metabolism. The dhfr and dhps gene mutations are known to be associated with sulphadoxine/ pyrimethamine (SP) resistance. The aim of this study is to investigate the single nucleotide polymorphisms (SNPs) associated with these mutations and to determine their nature and distributions. A total number of 390 pregnant women were recruited, 336 of them were taking SP while 54 of the pregnant women were using other anti-malaria drugs for treatment of malaria. The polymerase chain reaction (PCR) technique was used to characterize the specie of the isolated plasmodium while Sanger sequencing method of molecular genotyping was adopted for the subjection of the Plasmodium to resistance studies using dhfr and dhps genes to identify possible mutations. We observed that about 64.86% sites of dhfr gene and 91.9% sites of dhps gene are polymorphic. Nonsynonymous mutation and biallelic polymorphisms were associated with the dhfr gene, which causes resistance to SP by Plasmodium vivax, since such mutation affects the coding sequence.
Keywords: Bioinformatics, Computational Biology
Introduction
Malaria is a parasitic infectious disease caused by parasites of the genus Plasmodium and is transmitted by mosquitoes. Drug resistance is one of the greatest challenges of malaria control programmes. Sulphadoxine-Pyrimethamine (SP) resistance is linked to substitutions of amino acids in the enzymes dihydropteroate synthetase (DHPS) and dihydrofolate reductase (DHFR) in the folate biosynthetic pathway [1, 2]. Pyrimethamine targets the enzyme DHFR disrupting catalysis of the Nicotinamide adenine dinucleotide phosphate hydrogen (NADPH)-dependent reduction of 7, 8-dihydrofolate to 5, 6, 7, 8-tetrahydrofolate [3]. Sulfadoxine blocks the folate biosynthetic pathway at the DHPS level by disrupting the coupling of 7, 8-dihydroxymethylpterin pyrophosphate with para-amino benzoic acid (PABA) to yield 7,8-dihydropteroate [4]. Sulphadoxine-Pyrimethamine resistance is linked to point mutations in the parasite genome specifically the dihydrofolate reductase (DHPS) genes. Mutations in DHFR confer resistance to pyrimethamime while mutations in DHPS confer resistance to sulfadoxine and other sulpha drugs. There are variations in SP mutations, it may be single, double or triple: the more mutations, the stronger the resistance. In sub- Saharan Africa, the DHFR triple mutant (Asn-108 + Ile-51 + Arg-59) and DHPS double mutant (Gly-437 + Glu-540) have been strongly associated with potential resistance [5, 6]. In a study done in Lagos, Nigeria, the mutant strains of P. falciparum were present in up to 75.9% of the blood samples of pregnant women. In another study done in six districts in the general population of Zambia, there was indication of variation of rates of resistance to SP with mutated DHFR frequency ranging from 71 - 92% and 39 -71% frequency for the double mutant DHPS respectively [7]. In the present study, the dhfr and dhps genes in P. vivax isolates collected from pregnant women and cord blood of these women in Nnewi, Anambra State, Nigeria were analyzed using the conventional PCR- Sequencing approach. The purpose was to understand the molecular variation associated with the antimalarial resistance and to assess the possibility of looking for novel drugs to replace SP.
Materials and Methods
Sample Collection
Maternal and cord blood samples were collected from pregnant women who were on SP for IPT. For each blood sample 20μl of blood was spotted on a piece of 3mm filter paper (Whatman, Maidstone, UK) and air-dried. The filter papers were identified properly using participant’s identification numbers. The dried filter paper samples were stored in individual zipper plastic bags with dryer at – 200 C until DNA extraction. Ethical approval was obtained from Nnamdi Azikiwe University Teaching Hospital, Nnewi, Ethical committee (NAUTH/CS/66/VOL.11/158/2018/092) and authorization from Primary healthcare centres and maternity homes administrations. The women’s informed consent was obtained before collecting their blood samples. The inclusion criteria include consenting pregnant women aged 18 – 45 years while Sickle cell disease patients and HIV patients were excluded from the study. Of 390 pregnant women recruited, the number infected with malaria was 68(20.2%) as confirmed with molecular diagnosis (PCR) in 336 SP users. Microscopy revealed 73(21.7%) in 390 pregnant women that were recruited (336 SP users and 54 non-users of SP). Molecular diagnosis of malaria was done only in 336 SP users.
DNA Extraction and Purification from Dried Blood Spots
DNA extracted from bloodspots on filter papers using QIA amp DNA mini kit [8].
Polymerase Chain Reaction (PCR)
Quick load, one Taq, one step polymerase chain reaction was used. Quick load one step PCR master (2x) with catalog number NEB MO486S was purchased from Inqaba Biotech., Hart field; South Africa incorporated and was used according to manufacturer’s instructions. The malaria diagnosis was established with PCR only and P.falciparum and P.vivax were distinguished with molecular diagnosis (PCR) only.
Preparation of Agarose Gel
One-point zero percent Agarose gel (1.0%) was prepared by dissolving 1.0g in 100ml Tris EDTA Buffer. The mixture was then heated in a microwave for 5minutes to dissolve completely. It was then allowed to cool at 560C and 6ul of Ethidium bromide was added to it. The Agarose gel was poured into the electrophoresis chambers with gel comb, and allowed to solidify.
Electrophoresis
Five micro liters of the amplified PCR products was analyzed on 1.5% Agarose gel containing Ethidium bromide in Tris EDTA buffer. Electrophoresis was performed at 90v for 6ominutes. After electrophoresis the PCR products were visualized by Wealth Dolphin Doc UV transilluminator and photographed. Molecular weights were calculated using molecular weight standard of the maker.
Polymerase Chain Reaction Product Cleaning and Purification
The PCR products were cleaned using Exonuclease/Shrimp Alkaline Phosphatase. Purification was done with ABI V.3.1 Big dye kit according to manufacturer’s instructions. The labeled products were then cleaned with Zymo Seq clean-up kit.
Sequencing
The Ultra-pure DNA was sequenced with ABI3500XL analyzer with a 50 cm array, using POP7 at Inqaba Biotechnical Industries Ltd (Hatfield, South Africa). Sequences data generated were analyzed with Geneious version 9.0.5 and phylogenetic trees were constructed using neighbor joining. The sequences were subsequently deposited in the National Center for Biotechnology Information (NCBI) database with their corresponding accession IDs (Table 1).
| S/N | Gene | NCBI Accession ID |
|---|---|---|
| 1 | dhfr | MT577725 |
| 2 | dhps | MT577726 |
Table 1: Pharmacokinetics properties of the newly glycosylated antibiotics.
Sequence Alignment
A total of 411 sequences (genotypes) of each dhfr and dhps genes from plasmodium vivax were retrieved from Genbank using the basic local alignment search tool based on 100% similarity to each of the query sequences. Thereafter, the retrieved sequences were aligned using the Clustal W algorithm.
Extracting Snps From The Alignments
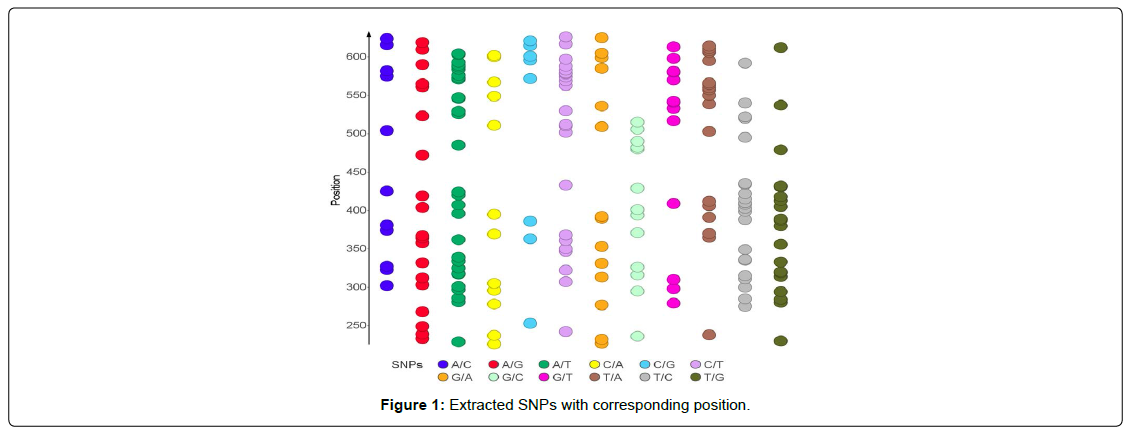
SNPs from the aligned sequences were extracted using adegenet package on R, which implements a parsimonious approach that allows for extracting SNPs from alignment while processing a reduced number of sequences at a time (Figure 1).
Result and Discussion
At regulatory sites, SNP rates are better and conservation is higher [9]. Within coding regions, SNP rates are highest and conservation is lowest at codon position 3 and the fewest SNPs are found at codon position two, reflecting codon degeneracy for amino acid encoding [10]. In this study, we found that DHFR and DHPS genes contains 24 and 12 SNPs respectively. In DHFR gene, the density of SNPS is higher at codon positions 1 and 2 and lowest at codon position 3 (Figure 2) while the level of SNPS in DHPS genes is highest at codon position 3 and almost equal at codon positions 1 and 2 (Figure 3). Single-nucleotide polymorphisms (SNPs) are considered to be the most common genetic changes that result from alterations in a single nucleotide [11]. Among SNPs, nonsynonymous SNPs (nsSNP) are associated with single amino acid substitution in the coding regions of a gene that may have the drastic effect on the structural and functional properties of the corresponding protein [12]. The second-codon position is the most functionally constrained; any change to the second codon position causes a nonsynonymous change in the coding sequence [13]. Because previous study showed that non-synonymous SNPs in K13 gene were found to be strongly associated with resistance to ART [14], therefore, we infer that the dominant presence of SNPs within the coding positions 1 and 2 in DHFR gene is associated with resistance to SP observed among the pregnant women, since dhfr mutations are known to be associated with sulphadoxine/pyrimethamine (SP) resistance [14].
In order to assess the rate and nature of polymorphism within the DHFR and DHPS genes, we observed that about 64.86% sites of DHFR gene and 91.9% sites of DHPS gene are polymorphic. This is not entirely surprising, given that the segments of both genes are known for their high mutation rate [15,16]. Considering the dhfr gene, all the polymorphic loci are biallelic (Figure 4) while the polymorphic sites in the dhps gene are 35.3%, 55.9% and 8.8% biallelic, triallelic and tetra-allelic respectively (Figure 5). Although most of the SNPs associated with human disease have been described as biallelic, in the last few years, an increasing number of these have been recognized to be triallelic and possibly even tetra-allelic [17].
While a large number of loci are nearly fixed (frequencies close to 0 or 1) in both DHFR and DHPS genes (Figure 6 and 7), there is an appreciable number of alleles with intermediate frequencies in dhps genes (Figure 6) and therefore susceptible to be associated with certain phenotypic traits.
Conclusion
The result revealed that although dhfr and dhps gene mutations are known to be associated with sulphadoxine/pyrimethamine (SP) resistance, the rate, nature and type of SNPs mutation differs between them. This gives further insight into the design and development of more potent antimalarials.
References
- Cowman AF, Mony MJ, Biggs BS, Cross GA, Foote SJ (1998) “Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthetase gene of Plasmodium falciparum dihydropterin pyrophosphokinase-dihydropteroate synthetase.” J of Bioorganic and Medi Chem 14:4433-4443.
- Brooks DR, Wang P, Read M, Watkins WM, Sims PF (1994) “Sequence variation of the hydroxymethyldihydropterin in pyrophosphokinase: dihydropteroate synthetase genes in lines of the human malaria parasite, Plasmodium falciparum with differing resistanceb to sulfadoxine.” Eur J of Biochem 224: 397 – 405.
- Walter RD (1991) “Folate metabolism as a target for chemotherapy of malaria.” In:coombs GH, North , MJ( edition). Biochemical Protozoology Tayor Francis, London: 560 – 568.
- Backeley RL (1984) “Distribution of dihydrofolate by dihydrofolate reductase.” J Biochemistry 23:2377 – 2383.
- Duraisingh MJ, Curtis J, Warhurst DS (1998) “Plasmodium falciparum detection of polymorphism in the dihydropfolate reductase and dihydropteroate synthetase genes by polymerase chain reaction and restriction digestion.” Experimental Parasitology 89:1-8.
- Triglia T, Menting JG, Wilson C, Cowman F (1997) “Mutations in dihydropteroate synthetase are responsible for sulphadoxine resistance in Plasmodium falciparum.” National Academy of Science 94:13944-13949
- Akanbi OM, Odaibo AB, Ademowo AG (2009) “The burden of malaria infection on pregnant women and birth weight of infants in Southern Western Nigeria.” East African J Pub Health 6: 63-68.
- Chandrasekhar BN, Jagannath M, Mulakkapurath NM, Sukriti M, Phani K.P et al. (2016) “Protocol for DNA Purification from Dried Blood Spots. Quality Improvement Agency amplification DNA mini and Blood mini Handbook, fifth edition, Public Library of Science.” San Francisco Califonia, United States of America 42 – 43.
- Bofkin, L, Goldman N (2007) “Variation in evolutionary processes at different codon positions.” Molecular Biology and Evolution 24: 513-521.
- Saleh Md (2016) “Impacts of nonsynonymous single nucleotide polymorphisms of adiponectin receptor 1 gene on corresponding protein stability: a computational approach." BioMed research international 2016.
- Dakal TC, Kala D, Dhiman G, Yadav V, Krokhotin A, et.al. (2017) “Predicting the functional consequences of non-synonymous single nucleotide polymorphisms in IL8 gene.” Scientific report 7: 1-18.
- Ghosh A, Saran N, Saha S (2020) “Survey of drug resistance associated gene mutations in Mycobacterium tuberculosis, ESKAPE and other bacterial species.” Scientific reports 10: 1-11.
- Zhao Y, Liu Z, Soe MT, Wang L, Soe TN et al. (2019) “Genetic variations associated with drug resistance markers in asymptomatic Plasmodium falciparum infections in Myanmar. Genes 10: 692.
- Hüebner C, Petermann I, Browning BL, Shelling AN, Ferguson LR. (2007) “Triallelic single nucleotide polymorphisms and genotyping error in genetic epidemiology studies: MDR1 (ABCB1) G2677/T/A as an example.” Cancer Epidemiology and Prevention Biomarkers 16: 1185-1192.
- Haque A, Sait KHW, Alam Q, Alam MZ, Anfinan N (2020) “MDR1 Gene Polymorphisms and Its Association With Expression as a Clinical Relevance in Terms of Response to Chemotherapy and Prognosis in Ovarian Cancer.” Frontiers in Genetics 11: 516.
- Jombart T, Collins C (2015) “Analysing genome-wide SNP data using adegenet.” 2:2.
- Quigley KM, Bay LK, van Oppen, MJ (2020) “Genome‐wide SNP analysis reveals an increase in adaptive genetic variation through selective breeding of coral.” Molecular Ecology 29: 2176-2188.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi